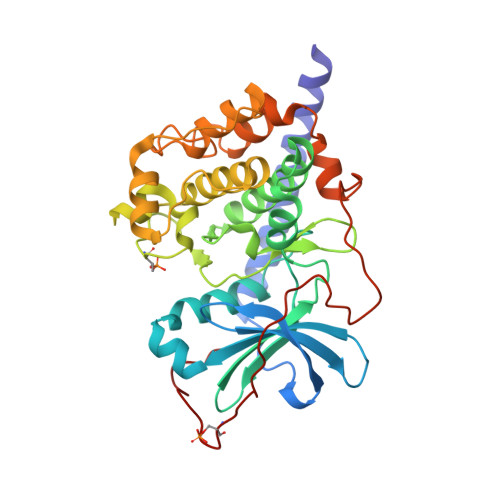
A conserved Glu-Arg salt bridge connects coevolved motifs that define the eukaryotic protein kinase fold.
Yang, J., Wu, J., Steichen, J.M., Kornev, A.P., Deal, M.S., Li, S., Sankaran, B., Woods, V.L., Taylor, S.S.(2012) J Mol Biology 415: 666-679
- PubMed: 22138346
- DOI: https://doi.org/10.1016/j.jmb.2011.11.035
- Primary Citation of Related Structures:
3QAL, 3QAM - PubMed Abstract:
Eukaryotic protein kinases (EPKs) feature two coevolved structural segments, the Activation segment, which starts with the Asp-Phe-Gly (DFG) and ends with the Ala-Pro-Glu (APE) motifs, and the helical GHI subdomain that comprises αG-αH-αI helices. Eukaryotic-like kinases have a much shorter Activation segment and lack the GHI subdomain. They thus lack the conserved salt bridge interaction between the APE Glu and an Arg from the GHI subdomain, a hallmark signature of EPKs. Although the conservation of this salt bridge in EPKs is well known and its implication in diseases has been illustrated by polymorphism analysis, its function has not been carefully studied. In this work, we use murine cAMP-dependent protein kinase (protein kinase A) as the model enzyme (Glu208 and Arg280) to examine the role of these two residues. We showed that Ala replacement of either residue caused a 40- to 120-fold decrease in catalytic efficiency of the enzyme due to an increase in K(m)(ATP) and a decrease in k(cat). Crystal structures, as well as solution studies, also demonstrate that this ion pair contributes to the hydrophobic network and stability of the enzyme. We show that mutation of either Glu or Arg to Ala renders both mutant proteins less effective substrates for upstream kinase phosphoinositide-dependent kinase 1. We propose that the Glu208-Arg280 pair serves as a center hub of connectivity between these two structurally conserved elements in EPKs. Mutations of either residue disrupt communication not only between the two segments but also within the rest of the molecule, leading to altered catalytic activity and enzyme regulation.
- Department of Chemistry and Biochemistry, University of California San Diego, San Diego, CA 92093, USA.
Organizational Affiliation: