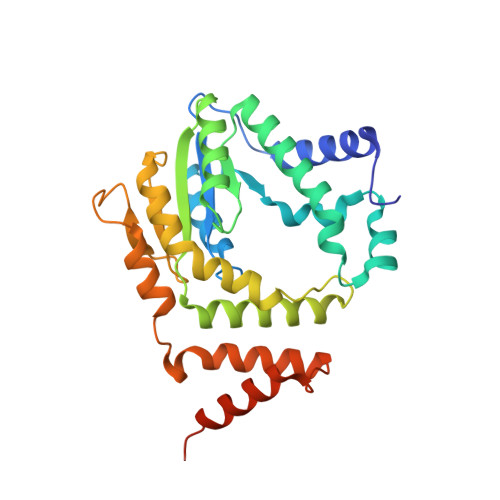
A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis.
Mutschler, H., Gebhardt, M., Shoeman, R.L., Meinhart, A.(2011) PLoS Biol 9: e1001033-e1001033
- PubMed: 21445328
- DOI: https://doi.org/10.1371/journal.pbio.1001033
- Primary Citation of Related Structures:
3Q8X - PubMed Abstract:
Most genomes of bacteria contain toxin-antitoxin (TA) systems. These gene systems encode a toxic protein and its cognate antitoxin. Upon antitoxin degradation, the toxin induces cell stasis or death. TA systems have been linked with numerous functions, including growth modulation, genome maintenance, and stress response. Members of the epsilon/zeta TA family are found throughout the genomes of pathogenic bacteria and were shown not only to stabilize resistance plasmids but also to promote virulence. The broad distribution of epsilon/zeta systems implies that zeta toxins utilize a ubiquitous bacteriotoxic mechanism. However, whereas all other TA families known to date poison macromolecules involved in translation or replication, the target of zeta toxins remained inscrutable. We used in vivo techniques such as microscropy and permeability assays to show that pneumococcal zeta toxin PezT impairs cell wall synthesis and triggers autolysis in Escherichia coli. Subsequently, we demonstrated in vitro that zeta toxins in general phosphorylate the ubiquitous peptidoglycan precursor uridine diphosphate-N-acetylglucosamine (UNAG) and that this activity is counteracted by binding of antitoxin. After identification of the product we verified the kinase activity in vivo by analyzing metabolite extracts of cells poisoned by PezT using high pressure liquid chromatograpy (HPLC). We further show that phosphorylated UNAG inhibitis MurA, the enzyme catalyzing the initial step in bacterial peptidoglycan biosynthesis. Additionally, we provide what is to our knowledge the first crystal structure of a zeta toxin bound to its substrate. We show that zeta toxins are novel kinases that poison bacteria through global inhibition of peptidoglycan synthesis. This provides a fundamental understanding of how epsilon/zeta TA systems stabilize mobile genetic elements. Additionally, our results imply a mechanism that connects activity of zeta toxin PezT to virulence of pneumococcal infections. Finally, we discuss how phosphorylated UNAG likely poisons additional pathways of bacterial cell wall synthesis, making it an attractive lead compound for development of new antibiotics.
- Department of Biomolecular Mechanisms, Max Planck Institute for Medical Research, Heidelberg, Germany.
Organizational Affiliation: