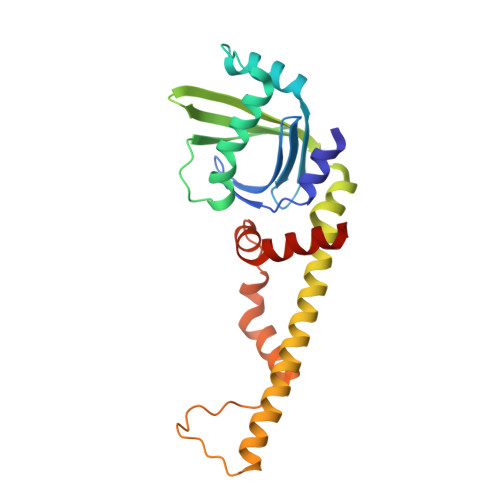
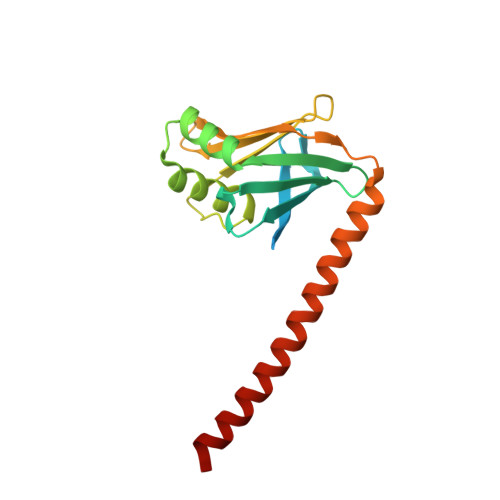
Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Ropars, V., Drevet, P., Legrand, P., Baconnais, S., Amram, J., Faure, G., Marquez, J.A., Pietrement, O., Guerois, R., Callebaut, I., Le Cam, E., Revy, P., de Villartay, J.P., Charbonnier, J.B.(2011) Proc Natl Acad Sci U S A 108: 12663-12668
- PubMed: 21768349
- DOI: https://doi.org/10.1073/pnas.1100758108
- Primary Citation of Related Structures:
3Q4F - PubMed Abstract:
Cernunnos/XLF is a core protein of the nonhomologous DNA end-joining (NHEJ) pathway that processes the majority of DNA double-strand breaks in mammals. Cernunnos stimulates the final ligation step catalyzed by the complex between DNA ligase IV and Xrcc4 (X4). Here we present the crystal structure of the X4(1-157)-Cernunnos(1-224) complex at 5.5-Å resolution and identify the relative positions of the two factors and their binding sites. The X-ray structure reveals a filament arrangement for X4(1-157) and Cernunnos(1-224) homodimers mediated by repeated interactions through their N-terminal head domains. A filament arrangement of the X4-Cernunnos complex was confirmed by transmission electron microscopy analyses both with truncated and full-length proteins. We further modeled the interface and used structure-based site-directed mutagenesis and calorimetry to characterize the roles of various residues at the X4-Cernunnos interface. We identified four X4 residues (Glu(55), Asp(58), Met(61), and Phe(106)) essential for the interaction with Cernunnos. These findings provide new insights into the molecular bases for stimulatory and bridging roles of Cernunnos in the final DNA ligation step.
- Laboratory of Structural Biology and Radiobiology, Commissariat à l'Energie Atomique et aux Energies Alternatives, Institut de Biologie et de Technologies de Saclay, 91191 Gif-s-Yvette, France.
Organizational Affiliation: