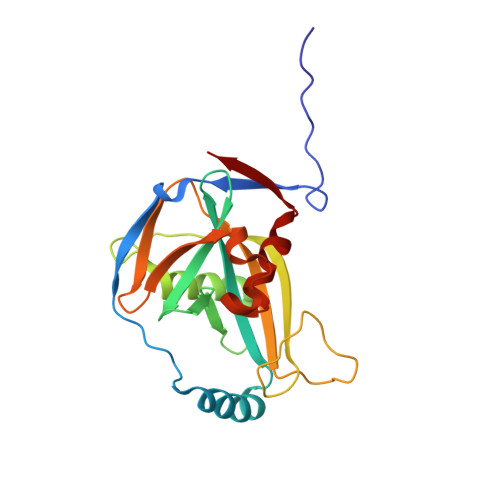
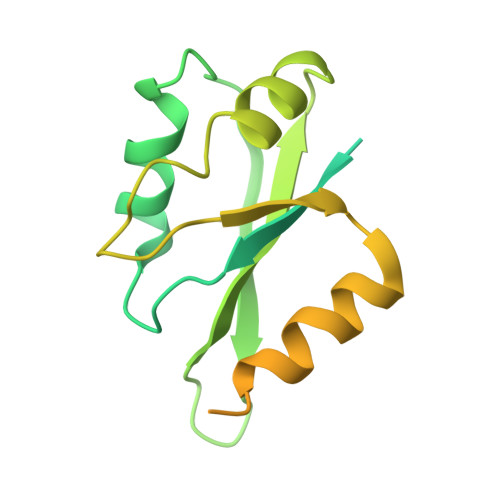
Crystal Structure of a Human Cleavage Factor CFI(m)25/CFI(m)68/RNA Complex Provides an Insight into Poly(A) Site Recognition and RNA Looping.
Yang, Q., Coseno, M., Gilmartin, G.M., Doublie, S.(2011) Structure 19: 368-377
- PubMed: 21295486
- DOI: https://doi.org/10.1016/j.str.2010.12.021
- Primary Citation of Related Structures:
3Q2S, 3Q2T - PubMed Abstract:
Cleavage factor I(m) (CFI(m)) is a highly conserved component of the eukaryotic mRNA 3' processing machinery that functions in sequence-specific poly(A) site recognition through the collaboration of a 25 kDa subunit containing a Nudix domain and a larger subunit of 59, 68, or 72 kDa containing an RNA recognition motif (RRM). Our previous work demonstrated that CFI(m)25 is both necessary and sufficient for sequence-specific binding of the poly(A) site upstream element UGUA. Here, we report the crystal structure of CFI(m)25 complexed with the RRM domain of CFI(m)68 and RNA. The CFI(m)25 dimer is clasped on opposite sides by two CFI(m)68 RRM domains. Each CFI(m)25 subunit binds one UGUA element specifically. Biochemical analysis indicates that the CFI(m)68 RRMs serve to enhance RNA binding and facilitate RNA looping. The intrinsic ability of CFI(m) to direct RNA looping may provide a mechanism for its function in the regulation of alternative poly(A) site selection.
- Department of Microbiology and Molecular Genetics, University of Vermont, Stafford Hall, Burlington, VT 05405, USA.
Organizational Affiliation: