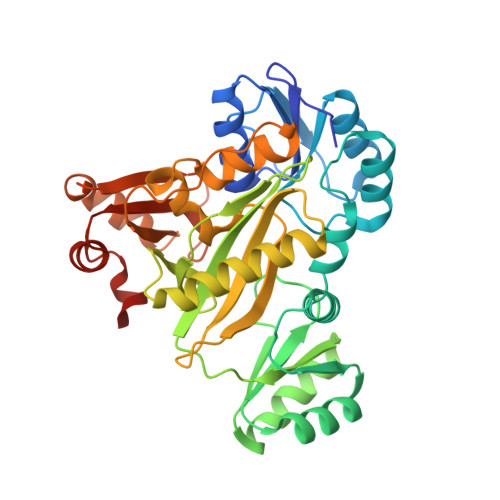
Structure of N(5)-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Tuntland, M.L., Johnson, M.E., Fung, L.W., Santarsiero, B.D.(2011) Acta Crystallogr D Biol Crystallogr 67: 870-874
- PubMed: 21931218
- DOI: https://doi.org/10.1107/S0907444911029210
- Primary Citation of Related Structures:
3Q2O - PubMed Abstract:
The apo structure of N5-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis (baPurK) with Mg2+ in the active site is reported at 1.96 Å resolution. PurK is an enzyme in the purine-biosynthetic pathway, unique to prokaryotes, that converts 5-aminoimidazole ribonucleotide to N5-carboxyaminoimidazole ribonucleotide and has been suggested as a potential antimicrobial drug target. Two interesting features of baPurK are a flexible B-loop (residues 149/150-157) that is in close contact with the active site and the binding of Mg2+ to the active site without additional ligands.
- Department of Chemistry, University of Illinois at Chicago, Chicago, IL 60607, USA.
Organizational Affiliation: