Structure-Based Design of a Protein Immunogen that Displays an HIV-1 gp41 Neutralizing Epitope.
Stanfield, R.L., Julien, J.P., Pejchal, R., Gach, J.S., Zwick, M.B., Wilson, I.A.(2011) J Mol Biology 414: 460-476
- PubMed: 22033480
- DOI: https://doi.org/10.1016/j.jmb.2011.10.014
- Primary Citation of Related Structures:
3Q1S - PubMed Abstract:
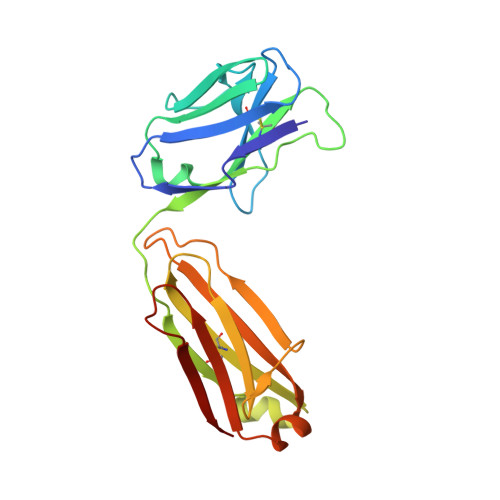
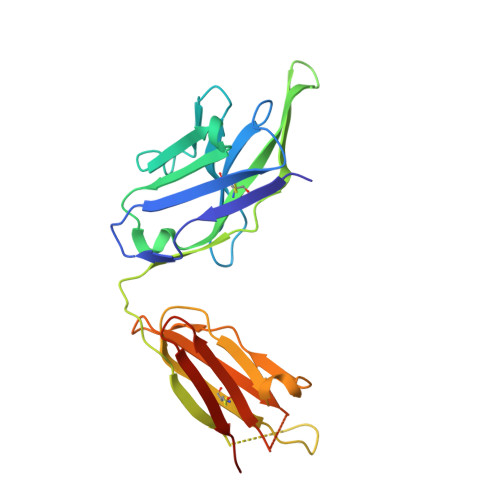
Antibody Z13e1 is a relatively broadly neutralizing anti-human immunodeficiency virus type 1 antibody that recognizes the membrane-proximal external region (MPER) of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Based on the crystal structure of an MPER epitope peptide in complex with Z13e1 Fab, we identified an unrelated protein, interleukin (IL)-22, with a surface-exposed region that is structurally homologous in its backbone to the gp41 Z13e1 epitope. By grafting the gp41 Z13e1 epitope sequence onto the structurally homologous region in IL-22, we engineered a novel protein (Z13-IL22-2) that contains the MPER epitope sequence for use as a potential immunogen and as a reagent for the detection of Z13e1-like antibodies. The Z13-IL22-2 protein binds Fab Z13e1 with a K(d) of 73 nM. The crystal structure of Z13-IL22-2 in complex with Fab Z13e1 shows that the epitope region is faithfully replicated in the Fab-bound scaffold protein; however, isothermal calorimetry studies indicate that Fab binding to Z13-IL22-2 is not a lock-and-key event, leaving open the question of whether conformational changes upon binding occur in the Fab, in Z13-IL-22, or in both.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA. robyn@scripps.edu
Organizational Affiliation: