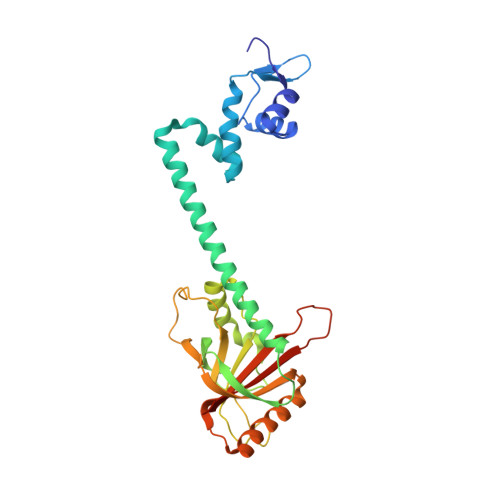
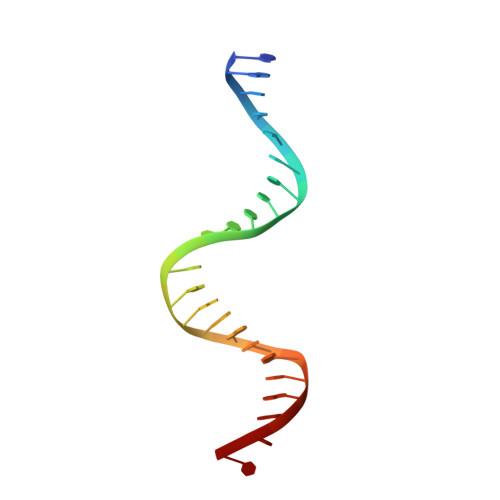
Structural contributions to multidrug recognition in the multidrug resistance (MDR) gene regulator, BmrR.
Bachas, S., Eginton, C., Gunio, D., Wade, H.(2011) Proc Natl Acad Sci U S A 108: 11046-11051
- PubMed: 21690368
- DOI: https://doi.org/10.1073/pnas.1104850108
- Primary Citation of Related Structures:
3Q1M, 3Q2Y, 3Q3D, 3Q5P, 3Q5R, 3Q5S - PubMed Abstract:
Current views of multidrug (MD) recognition focus on large drug-binding cavities with flexible elements. However, MD recognition in BmrR is supported by a small, rigid drug-binding pocket. Here, a detailed description of MD binding by the noncanonical BmrR protein is offered through the combined use of X-ray and solution studies. Low shape complementarity, suboptimal packing, and efficient burial of a diverse set of ligands is facilitated by an aromatic docking platform formed by a set of conformationally fixed aromatic residues, hydrophobic pincer pair that locks the different drug structures on the adaptable platform surface, and a trio of acidic residues that enables cation selectivity without much regard to ligand structure. Within the binding pocket is a set of BmrR-derived H-bonding donor and acceptors that solvate a wide range of ligand polar substituent arrangements in a manner analogous to aqueous solvent. Energetic analyses of MD binding by BmrR are consistent with structural data. A common binding orientation for the different BmrR ligands is in line with promiscuous allosteric regulation.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins School of Medicine, Baltimore, MD 21205, USA.
Organizational Affiliation: