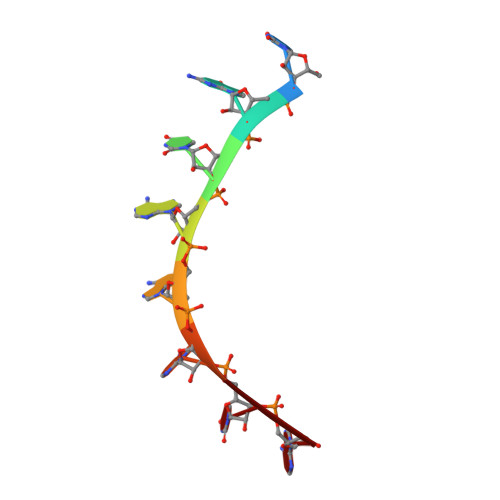
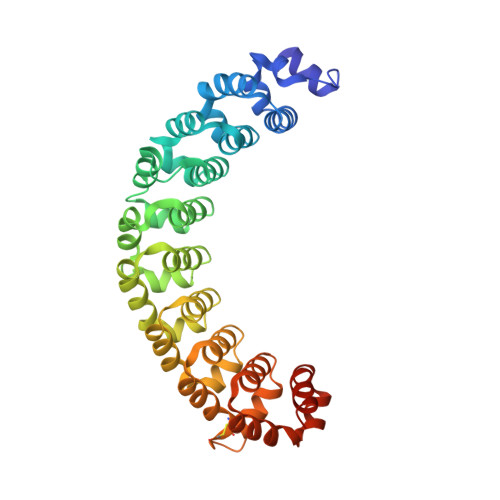Alternate modes of cognate RNA recognition by human PUMILIO proteins.
Lu, G., Hall, T.M.(2011) Structure 19: 361-367
- PubMed: 21397187
- DOI: https://doi.org/10.1016/j.str.2010.12.019
- Primary Citation of Related Structures:
3Q0L, 3Q0M, 3Q0N, 3Q0O, 3Q0P, 3Q0Q, 3Q0R, 3Q0S - PubMed Abstract:
Human PUMILIO1 (PUM1) and PUMILIO2 (PUM2) are members of the PUMILIO/FBF (PUF) family that regulate specific target mRNAs posttranscriptionally. Recent studies have identified mRNA targets associated with human PUM1 and PUM2. Here, we explore the structural basis of natural target RNA recognition by human PUF proteins through crystal structures of the RNA-binding domains of PUM1 and PUM2 in complex with four cognate RNA sequences, including sequences from p38α and erk2 MAP kinase mRNAs. We observe three distinct modes of RNA binding around the fifth RNA base, two of which are different from the prototypical 1 repeat:1 RNA base binding mode previously identified with model RNA sequences. RNA-binding affinities of PUM1 and PUM2 are not affected dramatically by the different binding modes in vitro. However, these modes of binding create structurally variable recognition surfaces that suggest a mechanism in vivo for recruitment of downstream effector proteins defined by the PUF:RNA complex.
- Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: