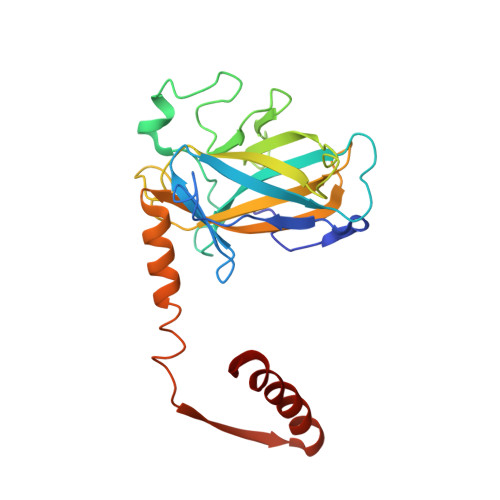
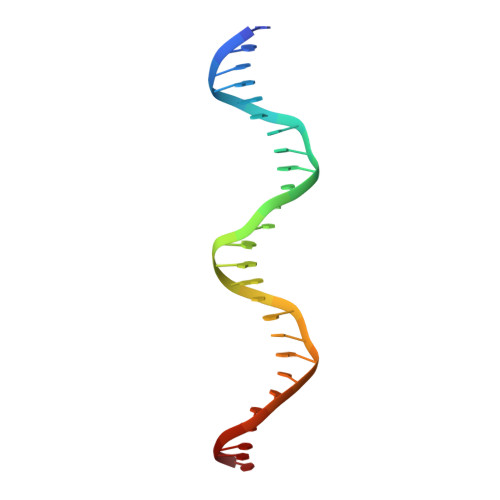
An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity.
Petty, T.J., Emamzadah, S., Costantino, L., Petkova, I., Stavridi, E.S., Saven, J.G., Vauthey, E., Halazonetis, T.D.(2011) EMBO J 30: 2167-2176
- PubMed: 21522129
- DOI: https://doi.org/10.1038/emboj.2011.127
- Primary Citation of Related Structures:
3Q01, 3Q05, 3Q06 - PubMed Abstract:
The p53 tumour suppressor gene, the most frequently mutated gene in human cancer, encodes a transcription factor that contains sequence-specific DNA binding and homo-tetramerization domains. Interestingly, the affinities of p53 for specific and non-specific DNA sites differ by only one order of magnitude, making it hard to understand how this protein recognizes its specific DNA targets in vivo. We describe here the structure of a p53 polypeptide containing both the DNA binding and oligomerization domains in complex with DNA. The structure reveals that sequence-specific DNA binding proceeds via an induced fit mechanism that involves a conformational switch in loop L1 of the p53 DNA binding domain. Analysis of loop L1 mutants demonstrated that the conformational switch allows DNA binding off-rates to be regulated independently of affinities. These results may explain the universal prevalence of conformational switching in sequence-specific DNA binding proteins and suggest that proteins like p53 rely more on differences in binding off-rates, than on differences in affinities, to recognize their specific DNA sites.
- Department of Molecular Biology, University of Geneva, Geneva, Switzerland.
Organizational Affiliation: