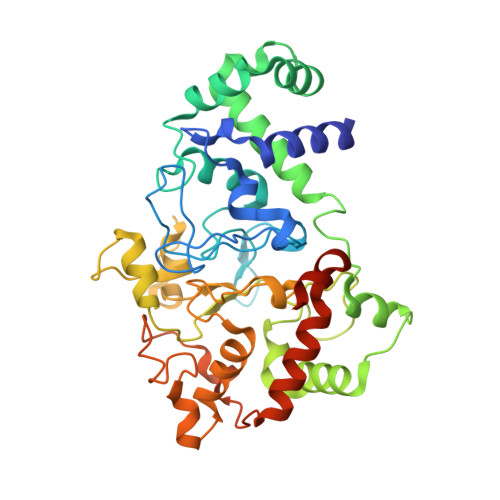
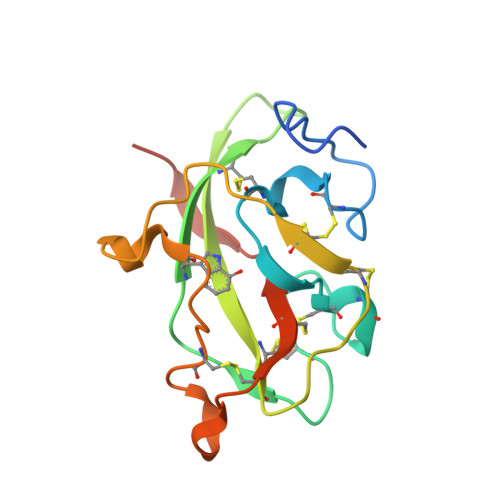
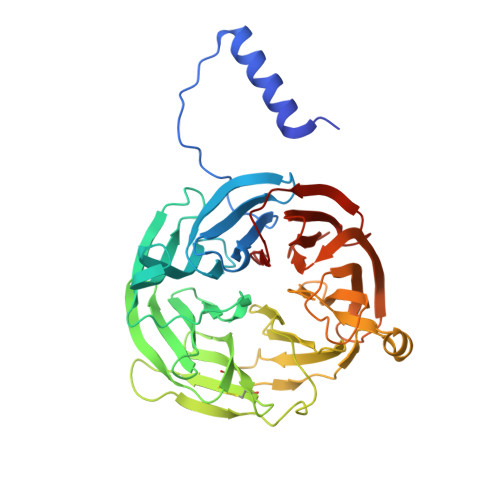
Crystal Structures of CO and NO Adducts of MauG in Complex with Pre-Methylamine Dehydrogenase: Implications for the Mechanism of Dioxygen Activation.
Yukl, E.T., Goblirsch, B.R., Davidson, V.L., Wilmot, C.M.(2011) Biochemistry 50: 2931-2938
- PubMed: 21355604
- DOI: https://doi.org/10.1021/bi200023n
- Primary Citation of Related Structures:
3PXS, 3PXT, 3PXW - PubMed Abstract:
MauG is a diheme enzyme responsible for the post-translational formation of the catalytic tryptophan tryptophylquinone (TTQ) cofactor in methylamine dehydrogenase (MADH). MauG can utilize hydrogen peroxide, or molecular oxygen and reducing equivalents, to complete this reaction via a catalytic bis-Fe(IV) intermediate. Crystal structures of diferrous, Fe(II)-CO, and Fe(II)-NO forms of MauG in complex with its preMADH substrate have been determined and compared to one another as well as to the structure of the resting diferric MauG-preMADH complex. CO and NO each bind exclusively to the 5-coordinate high-spin heme with no change in ligation of the 6-coordinate low-spin heme. These structures reveal likely roles for amino acid residues in the distal pocket of the high-spin heme in oxygen binding and activation. Glu113 is implicated in the protonation of heme-bound diatomic oxygen intermediates in promoting cleavage of the O-O bond. Pro107 is shown to change conformation on the binding of each ligand and may play a steric role in oxygen activation by positioning the distal oxygen near Glu113. Gln103 is in a position to provide a hydrogen bond to the Fe(IV)═O moiety that may account for the unusual stability of this species in MauG.
- Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, 321 Church Street SE, Minneapolis, Minnesota 55455, United States.
Organizational Affiliation: