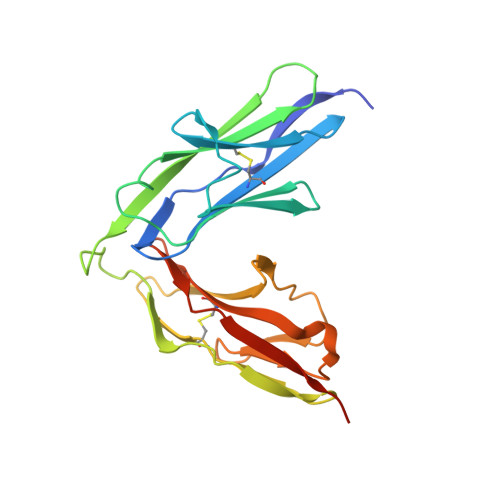
The Immunoglobulin-like Domains 1 and 2 of the Protein Tyrosine Phosphatase LAR Adopt an Unusual Horseshoe-like Conformation.
Biersmith, B.H., Hammel, M., Geisbrecht, E.R., Bouyain, S.(2011) J Mol Biology 408: 616-627
- PubMed: 21402080
- DOI: https://doi.org/10.1016/j.jmb.2011.03.013
- Primary Citation of Related Structures:
3PXH, 3PXJ - PubMed Abstract:
Neurogenesis depends on exquisitely regulated interactions between macromolecules on the cell surface and in the extracellular matrix. In particular, interactions between proteoglycans and members of the type IIa subgroup of receptor protein tyrosine phosphatases underlie crucial developmental processes such as the formation of synapses at the neuromuscular junction and the migration of axons to their appropriate targets. We report the crystal structures of the first and second immunoglobulin-like domains of the Drosophila type IIa receptor Dlar and its mouse homolog LAR. These two domains adopt an unusual antiparallel arrangement that has not been reported in tandem repeats of immunoglobulin-like domains and that is presumably conserved in all type IIa receptor protein tyrosine phosphatases.
- Division of Cell Biology and Biophysics, School of Biological Sciences, University of Missouri-Kansas City, 5100 Rockhill Road, Kansas City, MO 64110, USA.
Organizational Affiliation: