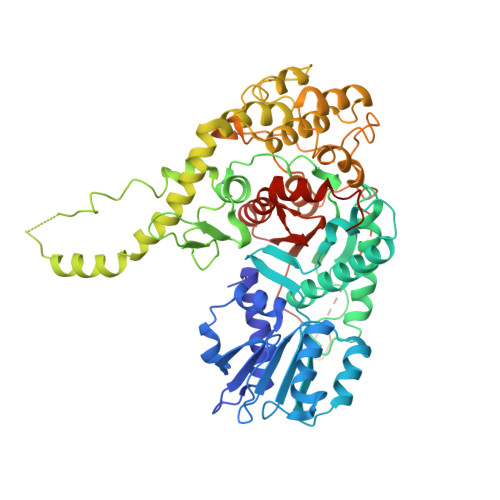
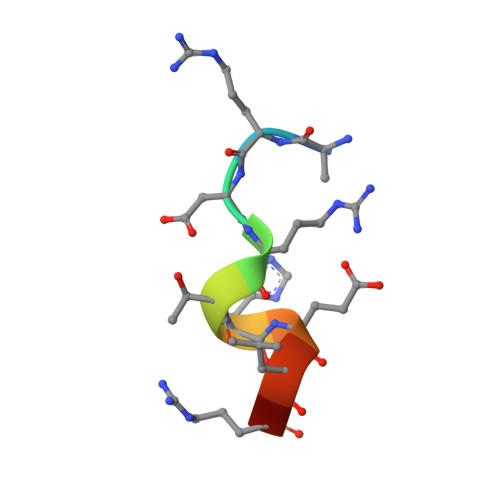
Possible roles for Munc18-1 domain 3a and Syntaxin1 N-peptide and C-terminal anchor in SNARE complex formation
Hu, S.-H., Christie, M.P., Saez, N.J., Latham, C.F., Jarrott, R., Lua, L.H.L., Collins, B.M., Martin, J.L.(2011) Proc Natl Acad Sci U S A 108: 1040-1045
- PubMed: 21193638
- DOI: https://doi.org/10.1073/pnas.0914906108
- Primary Citation of Related Structures:
3PUJ, 3PUK - PubMed Abstract:
Munc18-1 and Syntaxin1 are essential proteins for SNARE-mediated neurotransmission. Munc18-1 participates in synaptic vesicle fusion via dual roles: as a docking/chaperone protein by binding closed Syntaxin1, and as a fusion protein that binds SNARE complexes in a Syntaxin1 N-peptide dependent manner. The two roles are associated with a closed-open Syntaxin1 conformational transition. Here, we show that Syntaxin N-peptide binding to Munc18-1 is not highly selective, suggesting that other parts of the SNARE complex are involved in binding to Munc18-1. We also find that Syntaxin1, with an N peptide and a physically anchored C terminus, binds to Munc18-1 and that this complex can participate in SNARE complex formation. We report a Munc18-1-N-peptide crystal structure that, together with other data, reveals how Munc18-1 might transit from a conformation that binds closed Syntaxin1 to one that may be compatible with binding open Syntaxin1 and SNARE complexes. Our results suggest the possibility that structural transitions occur in both Munc18-1 and Syntaxin1 during their binary interaction. We hypothesize that Munc18-1 domain 3a undergoes a conformational change that may allow coiled-coil interactions with SNARE complexes.
- Institute for Molecular Bioscience, Division of Chemistry and Structural Biology, The University of Queensland, Brisbane, Queensland 4072, Australia.
Organizational Affiliation: