Design and synthesis of a novel, orally active, brain penetrant, tri-substituted thiophene based JNK inhibitor.
Bowers, S., Truong, A.P., Neitz, R.J., Neitzel, M., Probst, G.D., Hom, R.K., Peterson, B., Galemmo, R.A., Konradi, A.W., Sham, H.L., Toth, G., Pan, H., Yao, N., Artis, D.R., Brigham, E.F., Quinn, K.P., Sauer, J.M., Powell, K., Ruslim, L., Ren, Z., Bard, F., Yednock, T.A., Griswold-Prenner, I.(2011) Bioorg Med Chem Lett 21: 1838-1843
- PubMed: 21316234
- DOI: https://doi.org/10.1016/j.bmcl.2011.01.046
- Primary Citation of Related Structures:
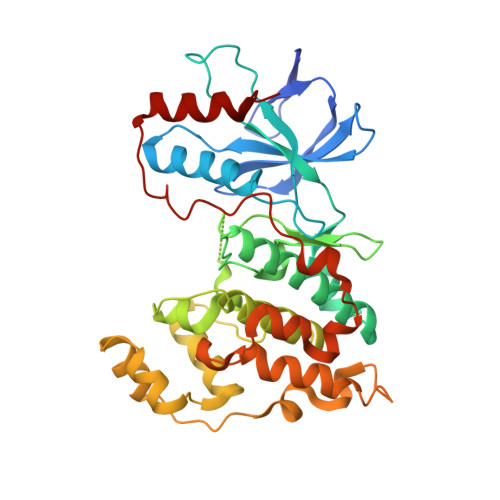
3PTG - PubMed Abstract:
The SAR of a series of tri-substituted thiophene JNK3 inhibitors is described. By optimizing both the N-aryl acetamide region of the inhibitor and the 4-position of the thiophene we obtained single digit nanomolar compounds, such as 47, which demonstrated an in vivo effect on JNK activity when dosed orally in our kainic acid mouse model as measured by phospho-c-jun reduction.
- Department of Chemical Sciences, Elan Pharmaceuticals, South San Francisco, CA 94080, USA. simeon.bowers@elan.com
Organizational Affiliation: