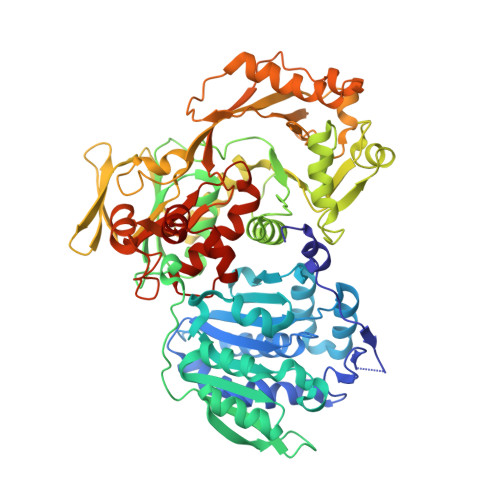
Structural basis for hypermodification of the wobble uridine in tRNA by bifunctional enzyme MnmC.
Kim, J., Almo, S.C.(2013) BMC Struct Biol 13: 5-5
- PubMed: 23617613
- DOI: https://doi.org/10.1186/1472-6807-13-5
- Primary Citation of Related Structures:
3PS9, 3PVC, 3SGL - PubMed Abstract:
Methylaminomethyl modification of uridine or 2-thiouridine (mnm5U34 or mnm5s2U34) at the wobble position of tRNAs specific for glutamate, lysine and arginine are observed in Escherichia coli and allow for specific recognition of codons ending in A or G. In the biosynthetic pathway responsible for this post-transcriptional modification, the bifunctional enzyme MnmC catalyzes the conversion of its hypermodified substrate carboxymethylaminomethyl uridine (cmnm5U34) to mnm5U34. MnmC catalyzes the flavin adenine dinucleotide (FAD)-dependent oxidative cleavage of carboxymethyl group from cmnm5U34 via an imine intermediate to generate aminomethyl uridine (nm5U34), which is subsequently methylated by S-adenosyl-L-methionine (SAM) to yield methylaminomethyl uridine (mnm5U34). The X-ray crystal structures of SAM/FAD-bound bifunctional MnmC from Escherichia coli and Yersinia pestis, and FAD-bound bifunctional MnmC from Yersinia pestis were determined and the catalytic functions verified in an in vitro assay. The crystal structures of MnmC from two Gram negative bacteria reveal the overall architecture of the enzyme and the relative disposition of the two independent catalytic domains: a Rossmann-fold domain containing the SAM binding site and an FAD containing domain structurally homologous to glycine oxidase from Bacillus subtilis. The structures of MnmC also reveal the detailed atomic interactions at the interdomain interface and provide spatial restraints relevant to the overall catalytic mechanism.
- Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, New York 10461, USA. jukim@aecom.yu.edu
Organizational Affiliation: