The crystal structure of the D307A mutant of alpha-Glucosidase (FAMILY 31) from Ruminococcus obeum ATCC 29174 in complex with acarbose
Tan, K., Tesar, C., Wilton, R., Keigher, L., Babnigg, G., Joachimiak, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| alpha-Glucosidase | 666 | Blautia obeum ATCC 29174 | Mutation(s): 1 Gene Names: RUMOBE_03919 EC: 3.2.1 | 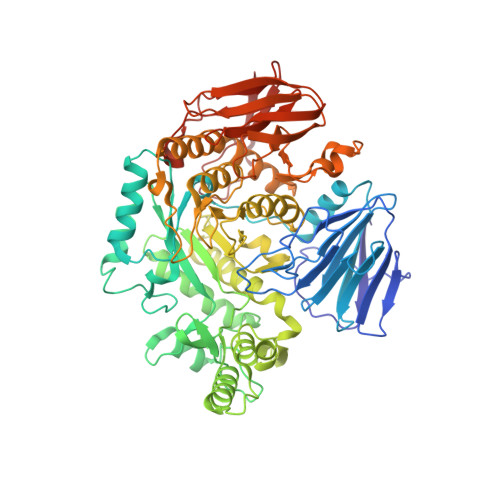 | |
UniProt | |||||
Find proteins for A5ZY13 (Blautia obeum ATCC 29174) Explore A5ZY13 Go to UniProtKB: A5ZY13 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A5ZY13 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | C | 3 | N/A | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G66431MI GlyCosmos: G66431MI | |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AC1 Query on AC1 | E [auth A], G [auth B] | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose C13 H23 N O8 RBZIIHWPZWOIDU-ZCGMLSCUSA-N |  | ||
| GLC Query on GLC | D [auth A] | alpha-D-glucopyranose C6 H12 O6 WQZGKKKJIJFFOK-DVKNGEFBSA-N |  | ||
| GOL Query on GOL | F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900007 Query on PRD_900007 | C | alpha-acarbose | Oligosaccharide / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.954 | α = 90 |
| b = 125.691 | β = 107.62 |
| c = 87.968 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |