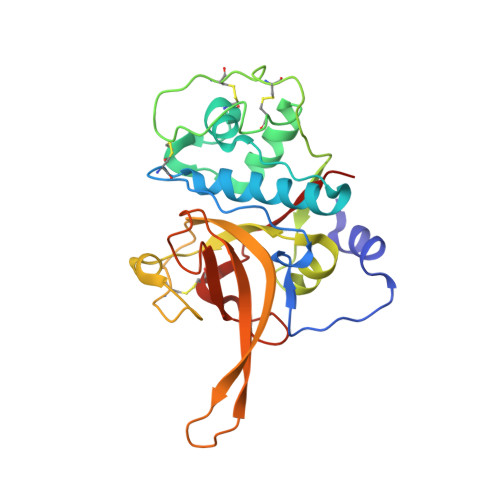
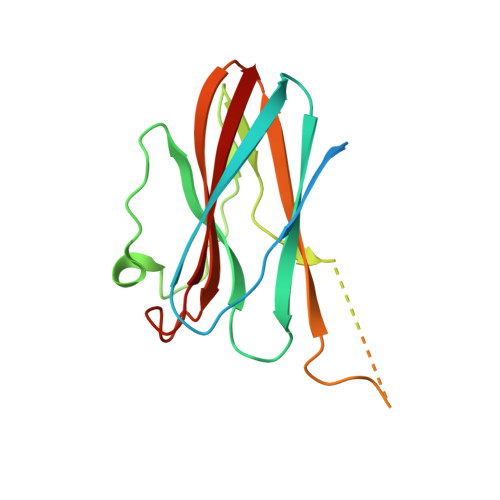
Structural basis for the regulation of cysteine-protease activity by a new class of protease inhibitors in Plasmodium.
Hansen, G., Heitmann, A., Witt, T., Li, H., Jiang, H., Shen, X., Heussler, V.T., Rennenberg, A., Hilgenfeld, R.(2011) Structure 19: 919-929
- PubMed: 21742259
- DOI: https://doi.org/10.1016/j.str.2011.03.025
- Primary Citation of Related Structures:
3PNR - PubMed Abstract:
Plasmodium cysteine proteases are essential for host-cell invasion and egress, hemoglobin degradation, and intracellular development of the parasite. The temporal, site-specific regulation of cysteine-protease activity is a prerequisite for survival and propagation of Plasmodium. Recently, a new family of inhibitors of cysteine proteases (ICPs) with homologs in at least eight Plasmodium species has been identified. Here, we report the 2.6 Å X-ray crystal structure of the C-terminal, inhibitory domain of ICP from P. berghei (PbICP-C) in a 1:1 complex with falcipain-2, an important hemoglobinase of Plasmodium. The structure establishes Plasmodium ICP as a member of the I42 class of chagasin-like protease inhibitors but with large insertions and differences in the binding mode relative to other family members. Furthermore, the PbICP-C structure explains why host-cell cathepsin B-like proteases and, most likely, also the protease-like domain of Plasmodium SERA5 (serine-repeat antigen 5) are no targets for ICP.
- Institute of Biochemistry, Center for Structural and Cell Biology in Medicine, University of Lübeck, 23538 Lübeck, Germany.
Organizational Affiliation: