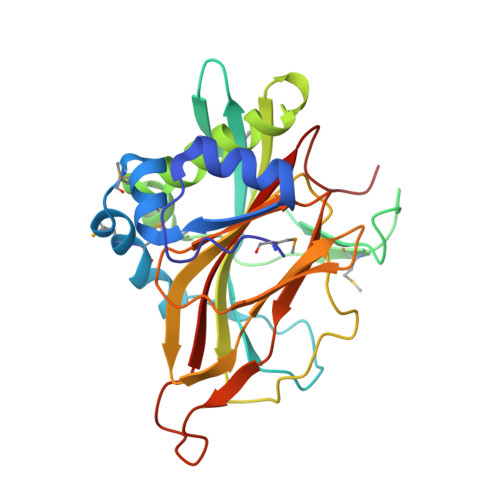
Crystal structure of a member of a novel family of dioxygenases (PF10014) reveals a conserved cupin fold and active site.
Xu, Q., Grant, J., Chiu, H.J., Farr, C.L., Jaroszewski, L., Knuth, M.W., Miller, M.D., Lesley, S.A., Godzik, A., Elsliger, M.A., Deacon, A.M., Wilson, I.A.(2014) Proteins 82: 164-170
- PubMed: 23852666
- DOI: https://doi.org/10.1002/prot.24362
- Primary Citation of Related Structures:
3PL0 - PubMed Abstract:
PF10014 is a novel family of 2-oxyglutarate-Fe(2+) -dependent dioxygenases that are involved in biosynthesis of antibiotics and regulation of biofilm formation, likely by catalyzing hydroxylation of free amino acids or other related ligands. The crystal structure of a PF10014 member from Methylibium petroleiphilum at 1.9 Å resolution shows strong structural similarity to cupin dioxygenases in overall fold and active site, despite very remote homology. However, one of the β-strands of the cupin catalytic core is replaced by a loop that displays conformational isomerism that likely regulates the active site.
- Joint Center for Structural Genomics, La Jolla, California (http://www.jcsg.org); Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park, California, 94025.
Organizational Affiliation: