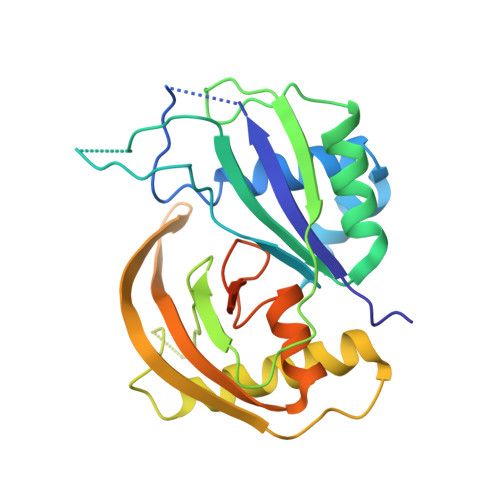
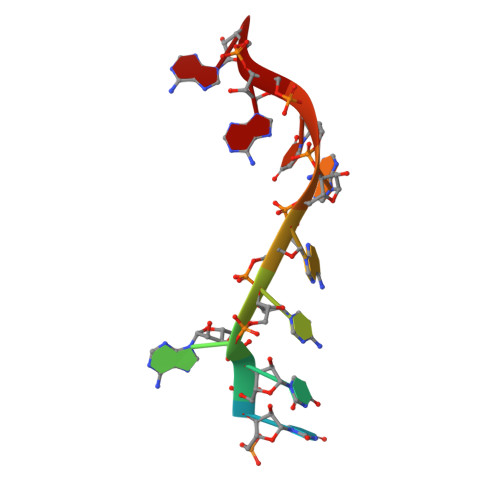
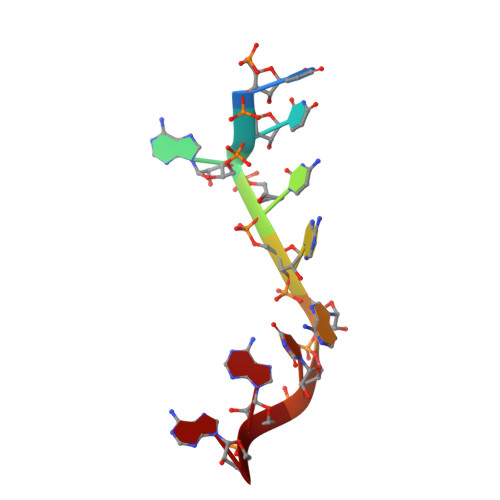
Interaction of the Cas6 Riboendonuclease with CRISPR RNAs: Recognition and Cleavage.
Wang, R., Preamplume, G., Terns, M.P., Terns, R.M., Li, H.(2011) Structure 19: 257-264
- PubMed: 21300293
- DOI: https://doi.org/10.1016/j.str.2010.11.014
- Primary Citation of Related Structures:
3PKM - PubMed Abstract:
The CRISPRs (Clustered Regularly Interspaced Short Palindromic Repeats) found in prokaryotic genomes confer small RNA-mediated protection against viruses and other invaders. CRISPR loci contain iterations of a short repeat sequence alternating with small segments of varying invader-derived sequences. Distinct families of CRISPR-associated Cas proteins function to cleave within the repeat sequence of CRISPR transcripts and produce the individual invader-targeting crRNAs. Here, we report the crystal structure of Pyrococcus furiosus Cas6 bound with a repeat RNA at 3.2 Å resolution. In contrast to other Cas families of endonucleases, Cas6 clasps nucleotides 2-9 of the repeat RNA using its two ferredoxin-like domains, and the enzyme-anchored 5' end tethers the distal cleavage site of the RNA between nucleotides 22 and 23 to the predicted enzyme active site on the opposite side of the ferrodoxin-like domains. Our findings suggest a wrap-around mechanism for CRISPR RNA recognition and cleavage by Cas6 and related processing endonucleases.
- Department of Chemistry and Biochemistry and Institute of Molecular Biophysics, Florida State University, Tallahassee, FL 32306, USA.
Organizational Affiliation: