Crystal Structures of Anopheles gambiae Odorant-binding Protein AgamOBP22a and Complexes with Bound Odorants
Ren, H., Yang, G., Winberg, G., Turin, L., Zhang, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Odorant binding protein (AGAP010409-PA) | 141 | Anopheles gambiae str. PEST | Mutation(s): 0 Gene Names: OBP22, OBPjj83b, AGAP010409 | 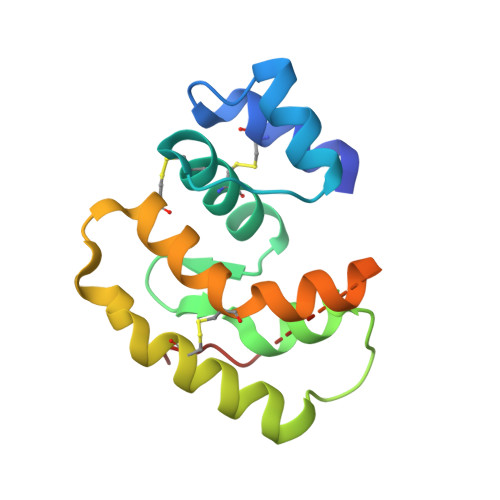 | |
UniProt | |||||
Find proteins for Q7PGA3 (Anopheles gambiae) Explore Q7PGA3 Go to UniProtKB: Q7PGA3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7PGA3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NH4 Query on NH4 | B [auth A], C [auth A], D [auth A] | AMMONIUM ION H4 N QGZKDVFQNNGYKY-UHFFFAOYSA-O |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 72.263 | α = 90 |
| b = 37.702 | β = 99.46 |
| c = 43.639 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CrystalClear | data collection |
| PHENIX | model building |
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHENIX | phasing |