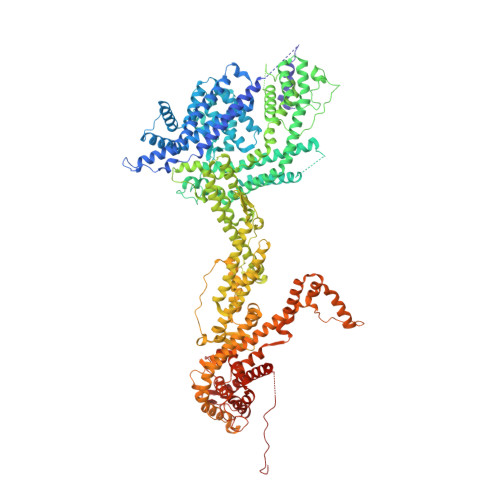
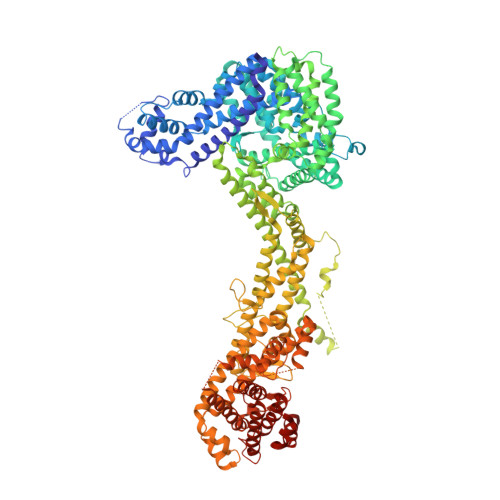
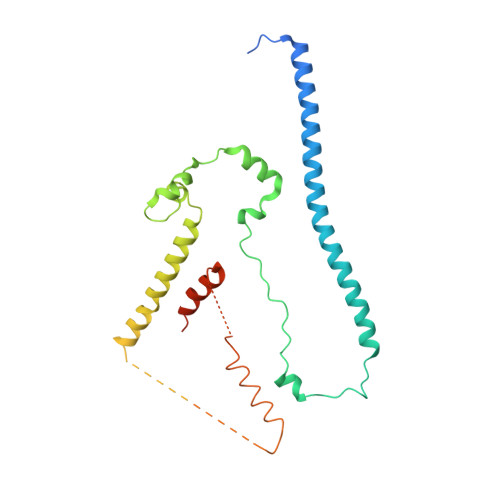
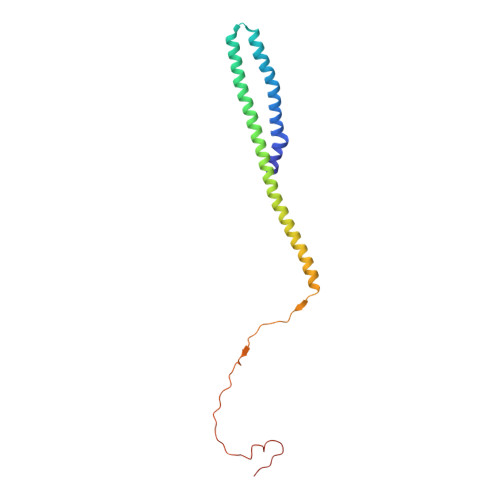
Structure and control of the actin regulatory WAVE complex.
Chen, Z., Borek, D., Padrick, S.B., Gomez, T.S., Metlagel, Z., Ismail, A.M., Umetani, J., Billadeau, D.D., Otwinowski, Z., Rosen, M.K.(2010) Nature 468: 533-538
- PubMed: 21107423
- DOI: https://doi.org/10.1038/nature09623
- Primary Citation of Related Structures:
3P8C - PubMed Abstract:
Members of the Wiskott-Aldrich syndrome protein (WASP) family control cytoskeletal dynamics by promoting actin filament nucleation with the Arp2/3 complex. The WASP relative WAVE regulates lamellipodia formation within a 400-kilodalton, hetero-pentameric WAVE regulatory complex (WRC). The WRC is inactive towards the Arp2/3 complex, but can be stimulated by the Rac GTPase, kinases and phosphatidylinositols. Here we report the 2.3-ångstrom crystal structure of the WRC and complementary mechanistic analyses. The structure shows that the activity-bearing VCA motif of WAVE is sequestered by a combination of intramolecular and intermolecular contacts within the WRC. Rac and kinases appear to destabilize a WRC element that is necessary for VCA sequestration, suggesting the way in which these signals stimulate WRC activity towards the Arp2/3 complex. The spatial proximity of the Rac binding site and the large basic surface of the WRC suggests how the GTPase and phospholipids could cooperatively recruit the complex to membranes.
- Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas, 5323 Harry Hines Boulevard, Dallas, Texas 75390, USA.
Organizational Affiliation: