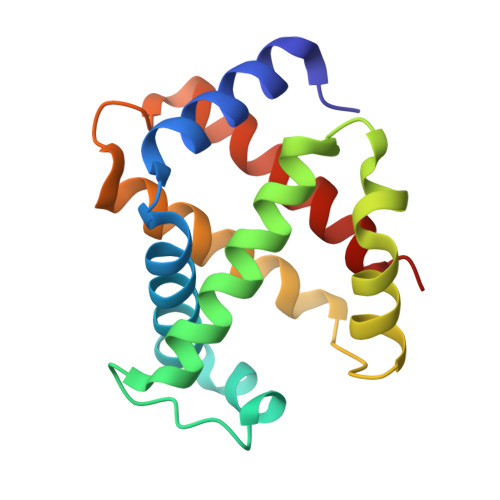
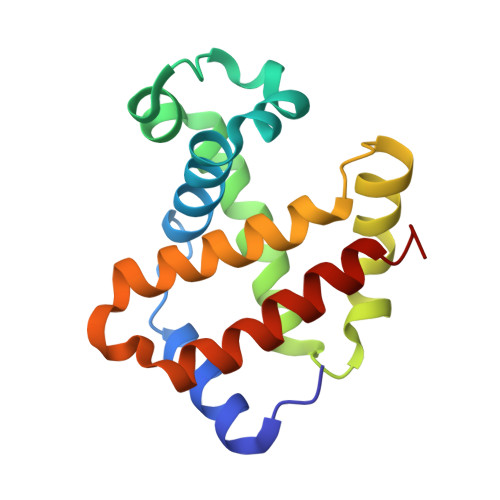
Structure of human R-state aquomethemoglobin at 2.0 A resolution
Yi, J., Thomas, L.M., Richter-Addo, G.B.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 647-651
- PubMed: 21636902
- DOI: https://doi.org/10.1107/S1744309111012528
- Primary Citation of Related Structures:
3P5Q - PubMed Abstract:
The crystal structure of tetrameric (αβ)(2) R-state human adult aquomethemoglobin is reported at 2.0 Å resolution. The asymmetric unit contained one αβ subunit pair. The R-state crystal belonged to space group P4(1)2(1)2, with unit-cell parameters a = b = 53.6, c = 192.8 Å. An Fe-bound water molecule was modeled into the heme distal pockets of each of the α and β subunits. In the α subunit, a highly ordered liganded water was modeled with an Fe-O(water) distance of 2.2 Å and appears to be protected against escape from the distal pocket by the conformation of the heme propionate groups, which point upwards towards the distal His58 residue aided by a hydrogen-bonding network involving the solvent. In the β subunit, the liganded water exhibited greater motion and was modeled with a longer Fe-O(water) distance of 2.5 Å; in this subunit both propionate groups point downwards away from the distal His63 residue, presumably allowing greater motion of the liganded water in and out of the distal pocket.
- Department of Chemistry and Biochemistry, University of Oklahoma, 101 Stephenson Parkway, Norman, OK 73019, USA. yijun@ou.edu
Organizational Affiliation: