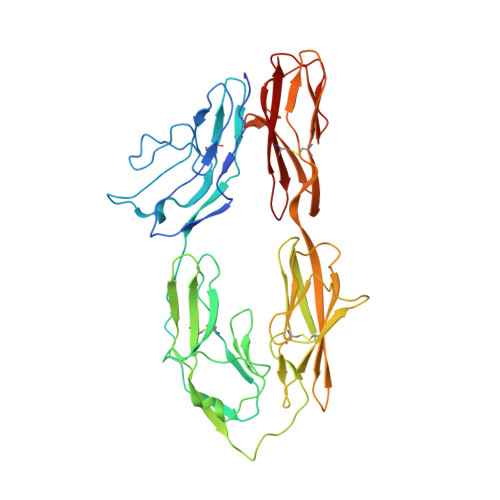
Homophilic adhesion mechanism of neurofascin, a member of the l1 family of neural cell adhesion molecules.
Liu, H., Focia, P.J., He, X.(2011) J Biological Chem 286: 797-805
- PubMed: 21047790
- DOI: https://doi.org/10.1074/jbc.M110.180281
- Primary Citation of Related Structures:
3P3Y, 3P40 - PubMed Abstract:
The L1 family neural cell adhesion molecules play key roles in specifying the formation and remodeling of the neural network, but their homophilic interaction that mediates adhesion is not well understood. We report two crystal structures of a dimeric form of the headpiece of neurofascin, an L1 family member. The four N-terminal Ig-like domains of neurofascin form a horseshoe shape, akin to several other immunoglobulin superfamily cell adhesion molecules such as hemolin, axonin, and Dscam. The neurofascin dimer, captured in two crystal forms with independent packing patterns, reveals a pair of horseshoes in trans-synaptic adhesion mode. The adhesion interaction is mediated mostly by the second Ig-like domain, which features an intermolecular β-sheet formed by the joining of two individual GFC β-sheets and a large but loosely packed hydrophobic cluster. Mutagenesis combined with gel filtration assays suggested that the side chain hydrogen bonds at the intermolecular β-sheet are essential for the homophilic interaction and that the residues at the hydrophobic cluster play supplementary roles. Our structures reveal a conserved homophilic adhesion mode for the L1 family and also shed light on how the pathological mutations of L1 affect its structure and function.
- Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA.
Organizational Affiliation: