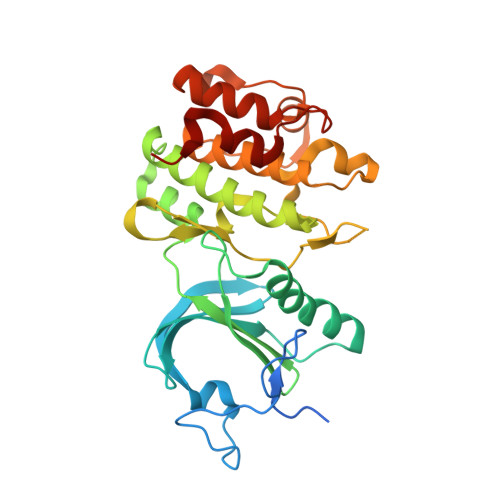
Structure of human Membrane-associated Tyrosine- and Threonine-specific cdc2-inhibitory kinase MYT1 (PKMYT1)
Chaikuad, A., Eswaran, J., Fedorov, O., Cooper, C.D.O., Kroeler, T., Vollmar, M., Krojer, T., Berridge, G., Muniz, J.R.C., Pike, A.C.W., von Delft, F., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S.To be published.