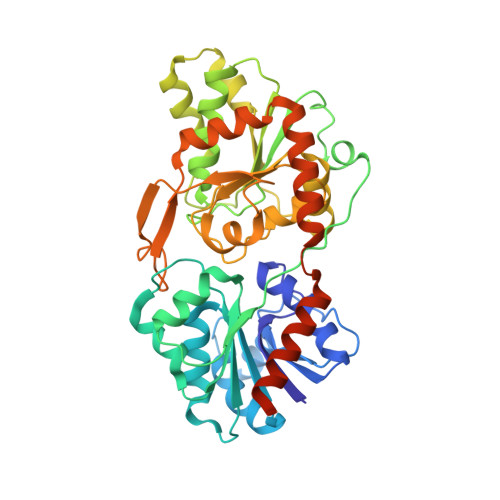
Crystal structure of a virus-encoded putative glycosyltransferase.
Xiang, Y., Baxa, U., Zhang, Y., Steven, A.C., Lewis, G.L., Van Etten, J.L., Rossmann, M.G.(2010) J Virol 84: 12265-12273
- PubMed: 20861263
- DOI: https://doi.org/10.1128/JVI.01303-10
- Primary Citation of Related Structures:
3OY2, 3OY7 - PubMed Abstract:
The chloroviruses (family Phycodnaviridae), unlike most viruses, encode some, if not most, of the enzymes involved in the glycosylation of their structural proteins. Annotation of the gene product B736L from chlorovirus NY-2A suggests that it is a glycosyltransferase. The structure of the recombinantly expressed B736L protein was determined by X-ray crystallography to 2.3-Å resolution, and the protein was shown to have two nucleotide-binding folds like other glycosyltransferase type B enzymes. This is the second structure of a chlorovirus-encoded glycosyltransferase and the first structure of a chlorovirus type B enzyme to be determined. B736L is a retaining enzyme and belongs to glycosyltransferase family 4. The donor substrate was identified as GDP-mannose by isothermal titration calorimetry and was shown to bind into the cleft between the two domains in the protein. The active form of the enzyme is probably a dimer in which the active centers are separated by about 40 Å.
- Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907-2054, USA.
Organizational Affiliation: