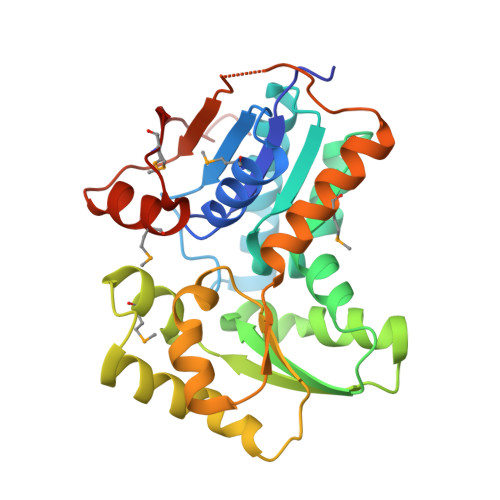
Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
Filippova, E.V., Wawrzak, Z., Onopriyenko, O., Kudriska, M., Edwards, A., Savchenko, A., Anderson, F.W., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.