Covalent Linkage of Distinct Substrate Degrons Controls Assembly and Disassembly of DegP Proteolytic Cages.
Kim, S., Grant, R.A., Sauer, R.T.(2011) Cell 145: 67-78
- PubMed: 21458668
- DOI: https://doi.org/10.1016/j.cell.2011.02.024
- Primary Citation of Related Structures:
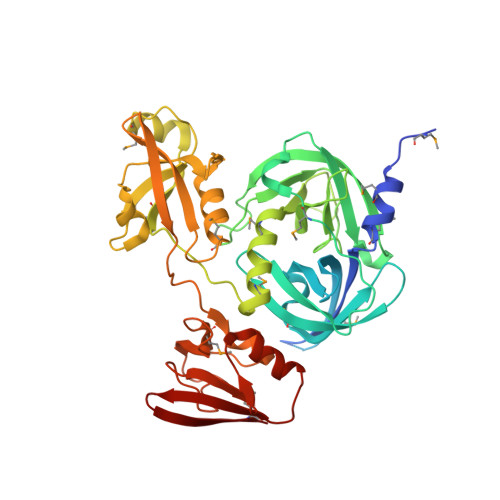
3OTP, 3OU0 - PubMed Abstract:
Protein quality control requires careful regulation of intracellular proteolysis. For DegP, a periplasmic protease, substrates promote assembly of inactive hexamers into proteolytically active cages with 12, 18, 24, or 30 subunits. Here, we show that sensitive activation and cage assembly require covalent linkage of distinct substrate sequences that affect degradation (degrons). One degron binds the DegP active site, and another degron binds a separate tethering site in PDZ1 in the crystal structure of a substrate-bound DegP dodecamer. FRET experiments demonstrate that active cages assemble rapidly in a reaction that is positively cooperative in substrate concentration, remain stably assembled while uncleaved substrate is present, and dissociate once degradation is complete. Thus, the energy of binding of linked substrate degrons drives assembly of the proteolytic machine responsible for subsequent degradation. Substrate cleavage and depletion results in disassembly, ensuring that DegP is proteolytically active only when sufficient quantities of protein substrates are present.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Organizational Affiliation: