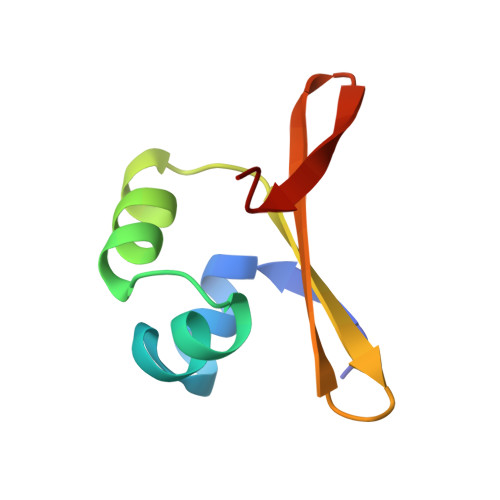
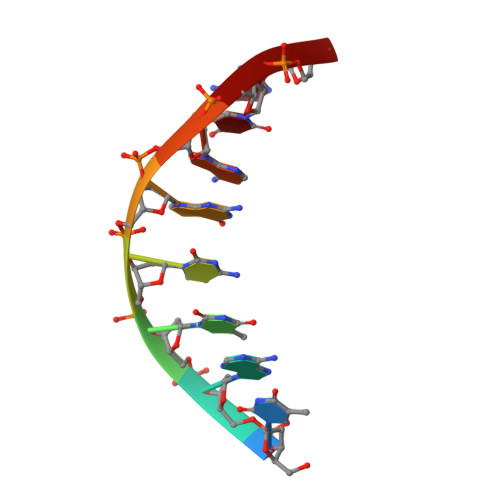
Crystal structure of an engineered Cro monomer bound nonspecifically to DNA: possible implications for nonspecific binding by the wild-type protein.
Albright, R.A., Mossing, M.C., Matthews, B.W.(1998) Protein Sci 7: 1485-1494
- PubMed: 9684880
- DOI: https://doi.org/10.1002/pro.5560070701
- Primary Citation of Related Structures:
3ORC - PubMed Abstract:
The structure has been determined at 3.0 A resolution of a complex of engineered monomeric Cro repressor with a seven-base pair DNA fragment. Although the sequence of the DNA corresponds to the consensus half-operator that is recognized by each subunit of the wild-type Cro dimer, the complex that is formed in the crystals by the isolated monomer appears to correspond to a sequence-independent mode of association. The overall orientation of the protein relative to the DNA is markedly different from that observed for Cro dimer bound to a consensus operator. The recognition helix is rotated 48 degrees further out of the major groove, while the turn region of the helix-turn-helix remains in contact with the DNA backbone. All of the direct base-specific interactions seen in the wild-type Cro-operator complex are lost. Virtually all of the ionic interactions with the DNA backbone, however, are maintained, as is the subset of contacts between the DNA backbone and a channel on the protein surface. Overall, 25% less surface area is buried at the protein DNA interface than for half of the wild-type Cro-operator complex, and the contacts are more ionic in character due to a reduction of hydrogen bonding and van der Waals interactions. Based on this crystal structure, model building was used to develop a possible model for the sequence-nonspecific interaction of the wild-type Cro dimer with DNA. In the sequence-specific complex, the DNA is bent, the protein dimer undergoes a large hinge-bending motion relative to the uncomplexed form, and the complex is twofold symmetric. In contrast, in the proposed nonspecific complex the DNA is straight, the protein retains a conformation similar to the apo form, and the complex lacks twofold symmetry. The model is consistent with thermodynamic, chemical, and mutagenic studies, and suggests that hinge bending of the Cro dimer may be critical in permitting the transition from the binding of protein at generic sites on the DNA to binding at high affinity operator sites.
- Howard Hughes Medical Institute and Department of Physics, University of Oregon, Eugene 97403, USA.
Organizational Affiliation: