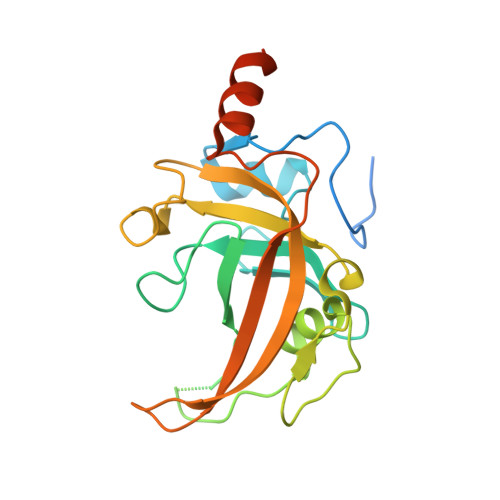
The Set and Ring Associated (SRA) domain of human ubiquitin-like containing PHD and RING finger domains 2 (UHRF2)
Walker, J.R., Xue, S., Avvakumov, G.V., Bountra, C., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bochkarev, A., Dhe-Paganon, S., Structural Genomics Consortium (SGC)To be published.