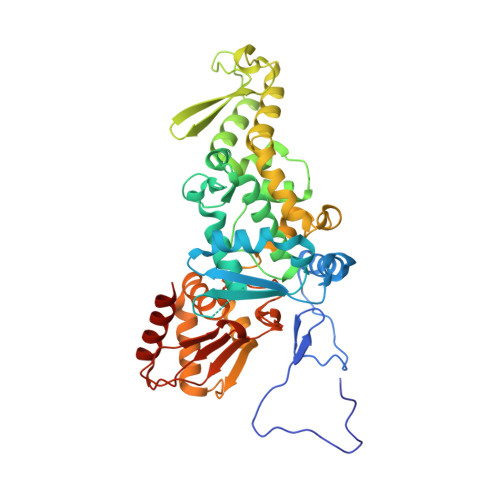
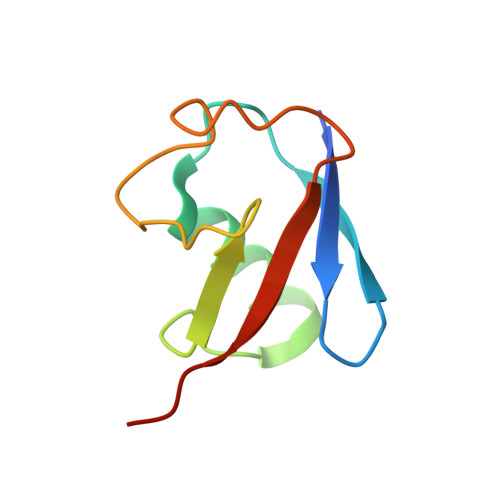
Structure and function of a HECT domain ubiquitin-binding site.
Kim, H.C., Steffen, A.M., Oldham, M.L., Chen, J., Huibregtse, J.M.(2011) EMBO Rep 12: 334-341
- PubMed: 21399621
- DOI: https://doi.org/10.1038/embor.2011.23
- Primary Citation of Related Structures:
3OLM - PubMed Abstract:
The Rsp5 ubiquitin ligase contains a non-covalent binding site for ubiquitin within the amino-terminal lobe (N-lobe) of the HECT domain, and the X-ray crystal structure of the HECT-ubiquitin complex has been determined. Hydrophobic patch residues of ubiquitin (L8, I44, V70) were crucial for interaction with Rsp5, and amino-acid alterations at the Rsp5-binding interface resulted in defects in polyubiquitination. Our results support a model in which the N-lobe-binding site acts to localize and orient the distal end of the ubiquitin chain to promote conjugation of the next ubiquitin molecule.
- Institute for Cellular and Molecular Biology, University of Texas, Austin, Texas 78712, USA.
Organizational Affiliation: