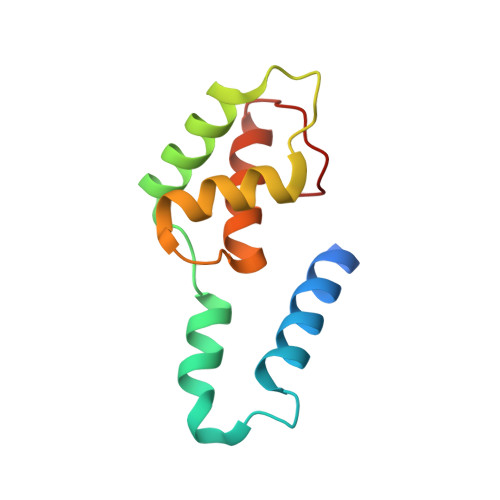
Structural diversity in the Mycobacteria DUF3349 superfamily.
Buchko, G.W., Abendroth, J., Robinson, J.I., Phan, I.Q., Myler, P.J., Edwards, T.E.(2020) Protein Sci 29: 670-685
- PubMed: 31658388
- DOI: https://doi.org/10.1002/pro.3758
- Primary Citation of Related Structures:
2LKY, 2M0N, 3OL3, 3OL4 - PubMed Abstract:
A protein superfamily with a "Domain of Unknown Function,", DUF3349 (PF11829), is present predominately in Mycobacterium and Rhodococcus bacterial species suggesting that these proteins may have a biological function unique to these bacteria. We previously reported the inaugural structure of a DUF3349 superfamily member, Mycobacterium tuberculosis Rv0543c. Here, we report the structures determined for three additional DUF3349 proteins: Mycobacterium smegmatis MSMEG_1063 and MSMEG_1066 and Mycobacterium abscessus MAB_3403c. Like Rv0543c, the NMR solution structure of MSMEG_1063 revealed a monomeric five α-helix bundle with a similar overall topology. Conversely, the crystal structure of MSMEG_1066 revealed a five α-helix protein with a strikingly different topology and a tetrameric quaternary structure that was confirmed by size exclusion chromatography. The NMR solution structure of a fourth member of the DUF3349 superfamily, MAB_3403c, with 18 residues missing at the N-terminus, revealed a monomeric α-helical protein with a folding topology similar to the three C-terminal helices in the protomer of the MSMEG_1066 tetramer. These structures, together with a GREMLIN-based bioinformatics analysis of the DUF3349 primary amino acid sequences, suggest two subfamilies within the DUF3349 family. The division of the DUF3349 into two distinct subfamilies would have been lost if structure solution had stopped with the first structure in the DUF3349 family, highlighting the insights generated by solving multiple structures within a protein superfamily. Future studies will determine if the structural diversity at the tertiary and quaternary levels in the DUF3349 protein superfamily have functional roles in Mycobacteria and Rhodococcus species with potential implications for structure-based drug discovery.
- Seattle Structural Genomics Center for Infectious Disease, Seattle, Washington.
Organizational Affiliation: