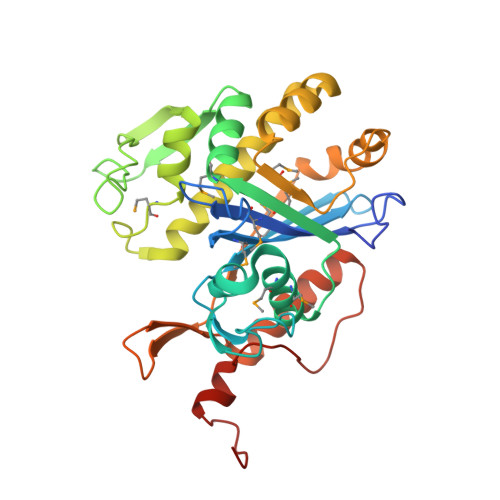
Northeast Structural Genomics Consortium Target SaR127
Kuzin, A., Lew, S., Vorobiev, S.M., Seetharaman, J., Janjua, J., Xiao, R., Ciccosanti, C., Wang, D., Everett, J.K., Nair, R., Acton, T.B., Rost, B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.