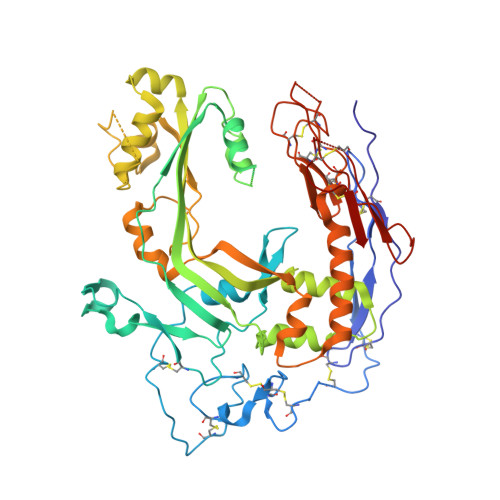
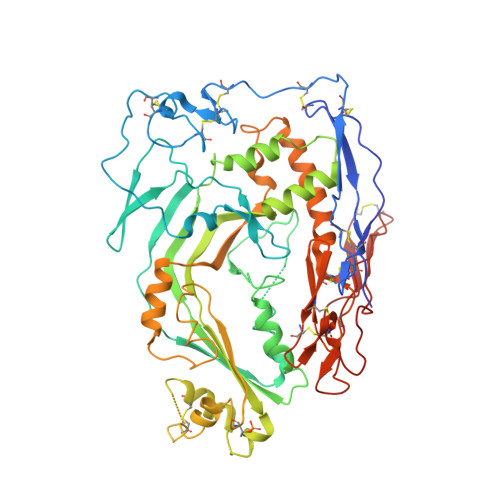
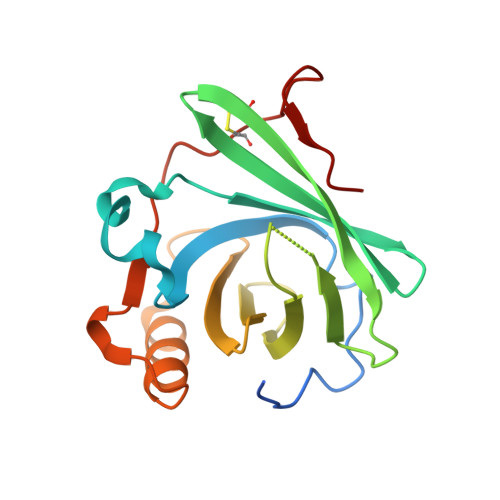
Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
Lovelace, L.L., Cooper, C.L., Sodetz, J.M., Lebioda, L.(2011) J Biological Chem 286: 17585-17592
- PubMed: 21454577
- DOI: https://doi.org/10.1074/jbc.M111.219766
- Primary Citation of Related Structures:
2RD7, 3OJY - PubMed Abstract:
C8 is one of five complement proteins that assemble on bacterial membranes to form the lethal pore-like "membrane attack complex" (MAC) of complement. The MAC consists of one C5b, C6, C7, and C8 and 12-18 molecules of C9. C8 is composed of three genetically distinct subunits, C8α, C8β, and C8γ. The C6, C7, C8α, C8β, and C9 proteins are homologous and together comprise the MAC family of proteins. All contain N- and C-terminal modules and a central 40-kDa membrane attack complex perforin (MACPF) domain that has a key role in forming the MAC pore. Here, we report the 2.5 Å resolution crystal structure of human C8 purified from blood. This is the first structure of a MAC family member and of a human MACPF-containing protein. The structure shows the modules in C8α and C8β are located on the periphery of C8 and not likely to interact with the target membrane. The C8γ subunit, a member of the lipocalin family of proteins that bind and transport small lipophilic molecules, shows no occupancy of its putative ligand-binding site. C8α and C8β are related by a rotation of ∼22° with only a small translational component along the rotation axis. Evolutionary arguments suggest the geometry of binding between these two subunits is similar to the arrangement of C9 molecules within the MAC pore. This leads to a model of the MAC that explains how C8-C9 and C9-C9 interactions could facilitate refolding and insertion of putative MACPF transmembrane β-hairpins to form a circular pore.
- Department of Chemistry and Biochemistry, University of South Carolina, Columbia, South Carolina 29208, USA.
Organizational Affiliation: