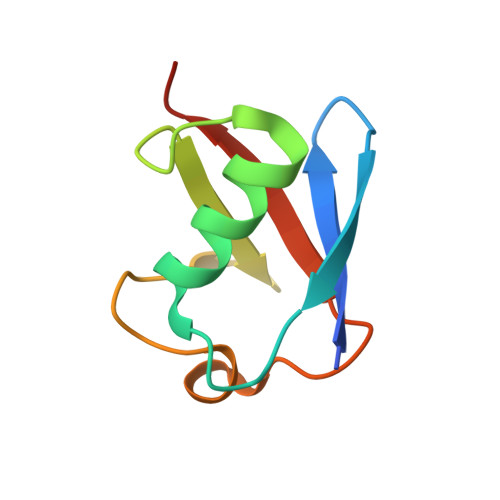
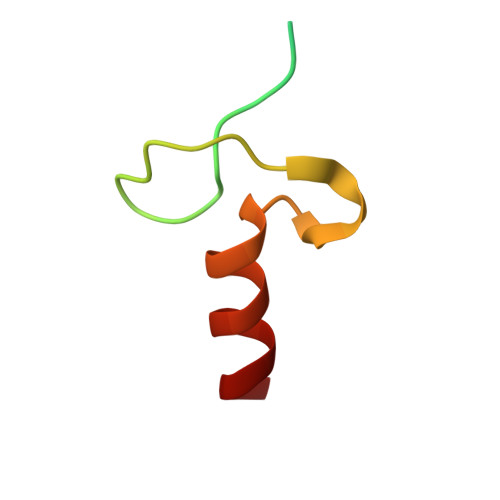
Ubiquitin Binding to A20 ZnF4 Is Required for Modulation of NF-KB Signaling
Bosanac, I., Wertz, I.E., Pan, B., Yu, C., Kusam, S., Lam, C., Phu, L., Phung, Q., Maurer, B., Arnott, D., Kirkpatrick, D.S., Dixit, V.M., Hymowitz, S.G.(2010) Mol Cell 40: 548-557
- PubMed: 21095585
- DOI: https://doi.org/10.1016/j.molcel.2010.10.009
- Primary Citation of Related Structures:
3OJ3, 3OJ4 - PubMed Abstract:
Inactivating mutations in the ubiquitin (Ub) editing protein A20 promote persistent nuclear factor (NF)-κB signaling and are genetically linked to inflammatory diseases and hematologic cancers. A20 tightly regulates NF-κB signaling by acting as an Ub editor, removing K63-linked Ub chains and mediating addition of Ub chains that target substrates for degradation. However, a precise molecular understanding of how A20 modulates this pathway remains elusive. Here, using structural analysis, domain mapping, and functional assays, we show that A20 zinc finger 4 (ZnF4) does not directly interact with E2 enzymes but instead can bind mono-Ub and K63-linked poly-Ub. Mutations to the A20 ZnF4 Ub-binding surface result in decreased A20-mediated ubiquitination and impaired regulation of NF-κB signaling. Collectively, our studies illuminate the mechanistically distinct but biologically interdependent activities of the A20 ZnF and ovarian tumor (OTU) domains that are inherent to the Ub editing process and, ultimately, to regulation of NF-κB signaling.
- Department of Structural Biology, Genentech, Inc, 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: