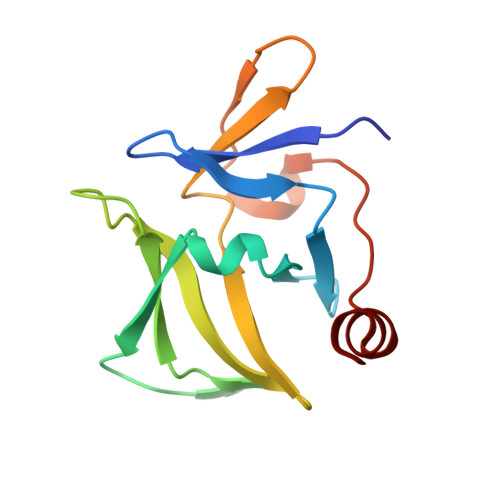
Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate.
Lu, C., Ranjith-Kumar, C.T., Hao, L., Kao, C.C., Li, P.(2011) Nucleic Acids Res 39: 1565-1575
- PubMed: 20961956
- DOI: https://doi.org/10.1093/nar/gkq974
- Primary Citation of Related Structures:
3OG8 - PubMed Abstract:
RIG-I recognizes molecular patterns in viral RNA to regulate the induction of type I interferons. The C-terminal domain (CTD) of RIG-I exhibits high affinity for 5' triphosphate (ppp) dsRNA as well as blunt-ended dsRNA. Structures of RIG-I CTD bound to 5'-ppp dsRNA showed that RIG-I recognizes the termini of dsRNA and interacts with the ppp through electrostatic interactions. However, the structural basis for the recognition of non-phosphorylated dsRNA by RIG-I is not fully understood. Here, we show that RIG-I CTD binds blunt-ended dsRNA in a different orientation compared to 5' ppp dsRNA and interacts with both strands of the dsRNA. Overlapping sets of residues are involved in the recognition of blunt-ended dsRNA and 5' ppp dsRNA. Mutations at the RNA-binding surface affect RNA binding and signaling by RIG-I. These results provide the mechanistic basis for how RIG-I recognizes different RNA ligands.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843-2128, USA.
Organizational Affiliation: