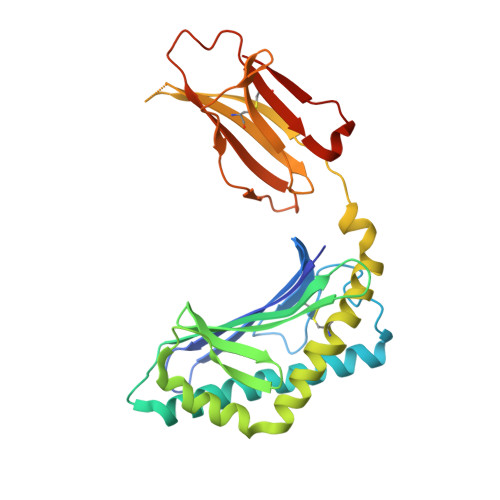
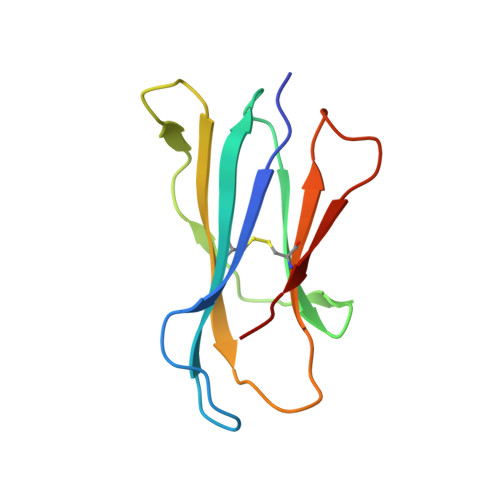
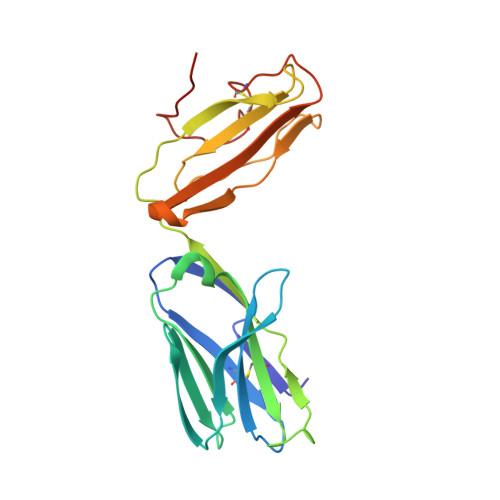
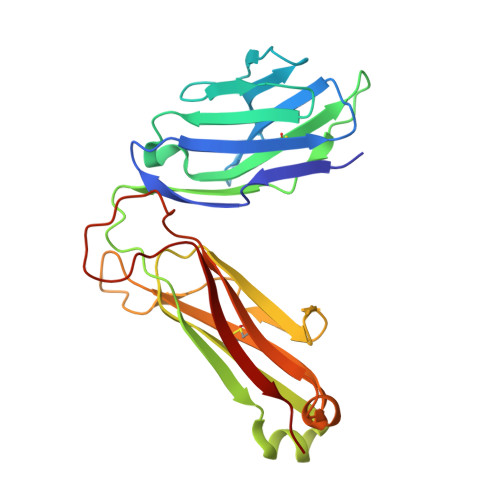
The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
Li, Y., Girardi, E., Wang, J., Yu, E.D., Painter, G.F., Kronenberg, M., Zajonc, D.M.(2010) J Exp Medicine 207: 2383-2393
- PubMed: 20921281
- DOI: https://doi.org/10.1084/jem.20101335
- Primary Citation of Related Structures:
3O8X, 3O9W - PubMed Abstract:
Invariant natural killer T cells (iNKT cells) rapidly produce effector cytokines. In this study, we report the first crystal structures of the iNKT cell T cell receptor (TCR) bound to two natural, microbial glycolipids presented by CD1d. Binding of the TCR induced CDR3-α-dependent structural changes in the F' roof of CD1d; these changes resemble those occurring in the absence of TCR engagement when the highly potent synthetic antigen α-galactosylceramide (α-GalCer) binds CD1d. Furthermore, in the Borrelia burgdorferi α-galactosyl diacylglycerol-CD1d complex, TCR binding caused a marked repositioning of the galactose sugar into an orientation that closely resembles α-GalCer. The TCR-dependent reorientation of the sugar, together with the induced CD1d fit, may explain the weaker potency of the microbial antigens compared with α-GalCer. We propose that the TCR of iNKT cells binds with a conserved footprint onto CD1d, regardless of the bound glycolipid antigen, and that for microbial antigens this unique binding mode requires TCR-initiated conformational changes.
- Division of Cell Biology, La Jolla Institute for Allergy and Immunology, La Jolla, CA 92037, USA.
Organizational Affiliation: