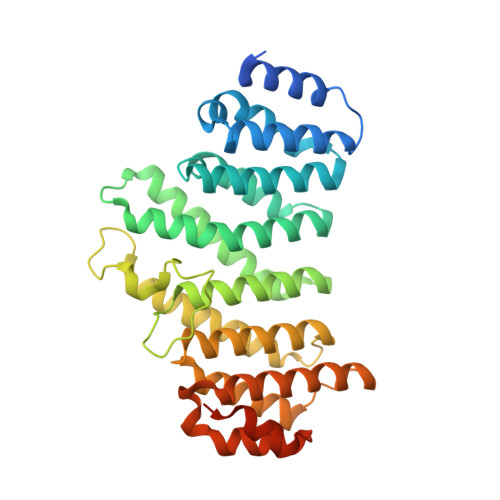
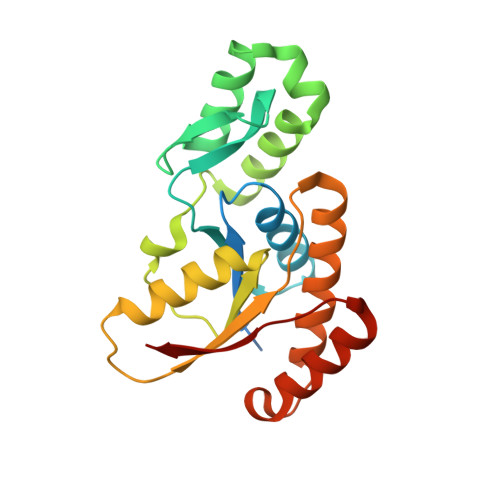
Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex.
Xiang, K., Nagaike, T., Xiang, S., Kilic, T., Beh, M.M., Manley, J.L., Tong, L.(2010) Nature 467: 729-733
- PubMed: 20861839
- DOI: https://doi.org/10.1038/nature09391
- Primary Citation of Related Structures:
3O2Q, 3O2S, 3O2T, 3ODR, 3ODS - PubMed Abstract:
Symplekin (Pta1 in yeast) is a scaffold in the large protein complex that is required for 3'-end cleavage and polyadenylation of eukaryotic messenger RNA precursors (pre-mRNAs); it also participates in transcription initiation and termination by RNA polymerase II (Pol II). Symplekin mediates interactions between many different proteins in this machinery, although the molecular basis for its function is not known. Here we report the crystal structure at 2.4 Å resolution of the amino-terminal domain (residues 30-340) of human symplekin in a ternary complex with the Pol II carboxy-terminal domain (CTD) Ser 5 phosphatase Ssu72 (refs 7, 10-17) and a CTD Ser 5 phosphopeptide. The N-terminal domain of symplekin has the ARM or HEAT fold, with seven pairs of antiparallel α-helices arranged in the shape of an arc. The structure of Ssu72 has some similarity to that of low-molecular-mass phosphotyrosine protein phosphatase, although Ssu72 has a unique active-site landscape as well as extra structural features at the C terminus that are important for interaction with symplekin. Ssu72 is bound to the concave face of symplekin, and engineered mutations in this interface can abolish interactions between the two proteins. The CTD peptide is bound in the active site of Ssu72, with the pSer 5-Pro 6 peptide bond in the cis configuration, which contrasts with all other known CTD peptide conformations. Although the active site of Ssu72 is about 25 Å from the interface with symplekin, we found that the symplekin N-terminal domain stimulates Ssu72 CTD phosphatase activity in vitro. Furthermore, the N-terminal domain of symplekin inhibits polyadenylation in vitro, but only when coupled to transcription. Because catalytically active Ssu72 overcomes this inhibition, our results show a role for mammalian Ssu72 in transcription-coupled pre-mRNA 3'-end processing.
- Department of Biological Sciences, Columbia University, New York, New York 10027, USA.
Organizational Affiliation: