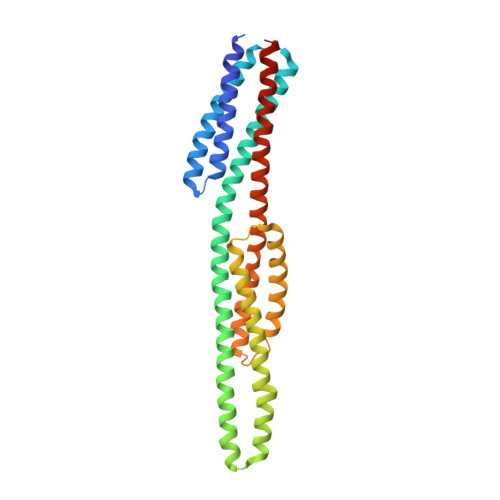
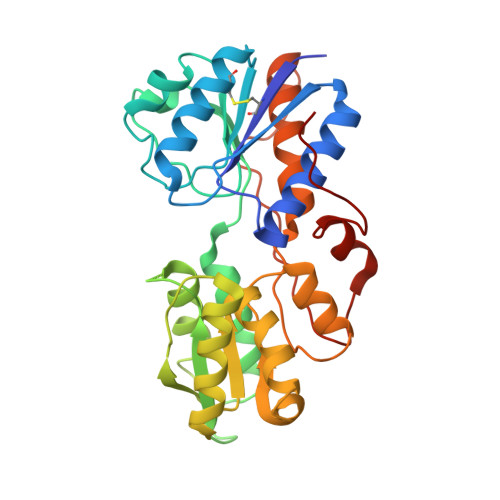
An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO.
Moore, J.O., Hendrickson, W.A.(2012) Structure 20: 729-741
- PubMed: 22483119
- DOI: https://doi.org/10.1016/j.str.2012.02.021
- Primary Citation of Related Structures:
3O1H, 3O1I, 3O1J - PubMed Abstract:
The osmoregulator trimethylamine-N-oxide (TMAO), commonplace in aquatic organisms, is used as the terminal electron acceptor for respiration in many bacterial species. The TMAO reductase (Tor) pathway for respiratory catalysis is controlled by a receptor system that comprises the TMAO-binding protein TorT, the sensor histidine kinase TorS, and the response regulator TorR. Here we study the TorS/TorT sensor system to gain mechanistic insight into signaling by histidine kinase receptors. We determined crystal structures for complexes of TorS sensor domains with apo TorT and with TorT (TMAO); we characterized TorS sensor associations with TorT in solution; we analyzed the thermodynamics of TMAO binding to TorT-TorS complexes; and we analyzed in vivo responses to TMAO through the TorT/TorS/TorR system to test structure-inspired hypotheses. TorS-TorT(apo) is an asymmetric 2:2 complex that binds TMAO with negative cooperativity to form a symmetric active kinase.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA.
Organizational Affiliation: