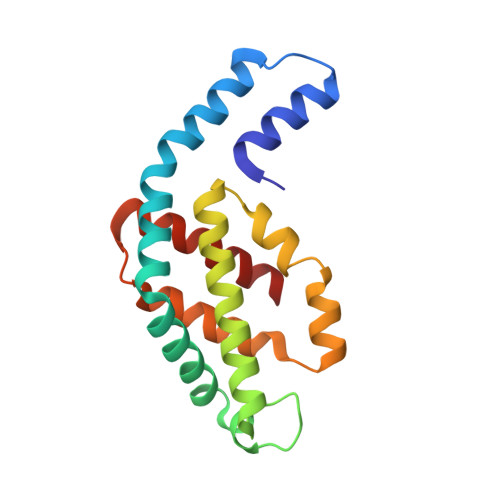
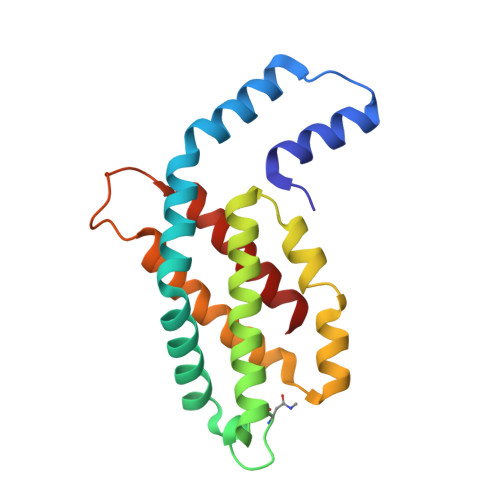
High-resolution crystal structures of trimeric and rod phycocyanin.
David, L., Marx, A., Adir, N.(2011) J Mol Biology 405: 201-213
- PubMed: 21035460
- DOI: https://doi.org/10.1016/j.jmb.2010.10.036
- Primary Citation of Related Structures:
3O18, 3O2C - PubMed Abstract:
The phycobilisome light-harvesting antenna in cyanobacteria and red algae is assembled from two substructures: a central core composed of allophycocyanin surrounded by rods that always contain phycocyanin (PC). Unpigmented proteins called linkers are also found within the rods and core. We present here two new structures of PC from the thermophilic cyanobacterium Thermosynechococcus vulcanus. We have determined the structure of trimeric PC to 1.35 Å, the highest resolution reported to date for this protein. We also present a structure of PC isolated in its intact and functional rod form at 1.5 Å. Analysis of rod crystals showed that in addition to the α and β PC subunit, there were three linker proteins: the capping rod linker (L(R)(8.7)), the rod linker (L(R)), and only one of three rod-core linkers (L(RC), CpcG4) with a stoichiometry of 12:12:1:1:1. This ratio indicates that the crystals contained rods composed of two hexamers. The crystallographic parameters of the rod crystals are nearly identical with that of the trimeric form, indicating that the linkers do not affect crystal packing and are completely embedded within the rod cavities. Absorption and fluorescence emission spectra were red-shifted, as expected for assembled rods, and this could be shown for the rod in solution as well as in crystal using confocal fluorescence microscopy. The crystal packing imparts superimposition of the three rod linkers, canceling out their electron density. However, analysis of B-factors and the conformations of residues facing the rod channel indicate the presence of linkers. Based on the experimental evidence presented here and a homology-based model of the L(R) protein, we suggest that the linkers do not in fact link between rod hexamers but stabilize the hexameric assembly and modify rod energy absorption and transfer capabilities.
- Schulich Faculty of Chemistry, Israel Institute of Technology, Haifa, 32000 Israel.
Organizational Affiliation: