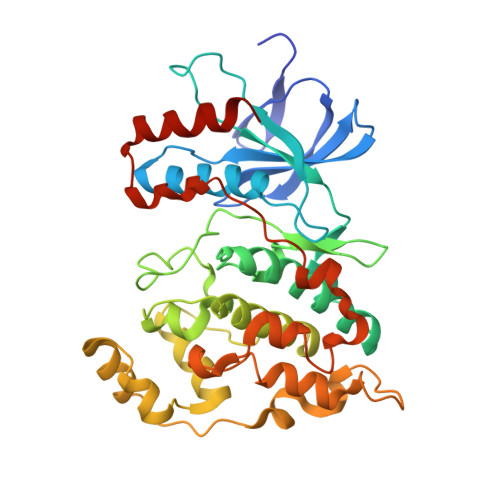
Crystal Structure of JNK1-alpha1 isoform
Comess, K.M., Sun, C., Abad-Zapatero, C., Borhani, D., Goedken, E., Argiriardi, M., Groebe, D.R., Gum, R.J., Jia, Y., Clampit, J.E., Haasch, D.L., Smith, H.T., Wang, S., Song, D., Coen, M.L., Cloutier, T.E., Tang, H., Cheng, X., Quinn, C., Liu, B., Xin, Z., Liu, G., Fry, E.H., Stoll, V., Ng, T.I., Banach, D., Marcotte, D., Burns, D.J., Hajduk, P.J.To be published.