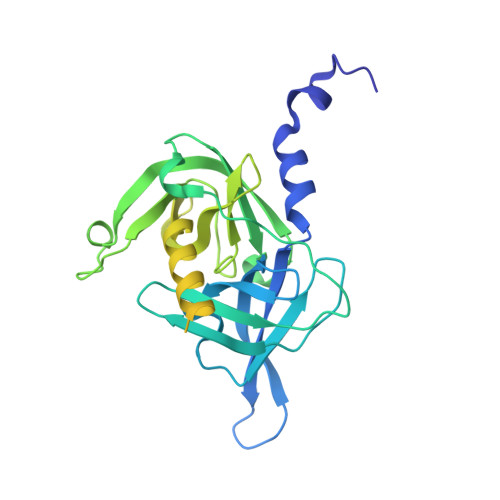
Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Truebestein, L., Tennstaedt, A., Monig, T., Krojer, T., Canellas, F., Kaiser, M., Clausen, T., Ehrmann, M.(2011) Nat Struct Mol Biol 18: 386-388
- PubMed: 21297635
- DOI: https://doi.org/10.1038/nsmb.2013
- Primary Citation of Related Structures:
3NUM, 3NWU, 3NZI - PubMed Abstract:
Crystal structures of active and inactive conformations of the human serine protease HTRA1 reveal that substrate binding to the active site is sufficient to stimulate proteolytic activity. HTRA1 attaches to liposomes, digests misfolded proteins into defined fragments and undergoes substrate-mediated oligomer conversion. In contrast to those of other serine proteases, the PDZ domain of HTRA1 is dispensable for activation or lipid attachment, indicative of different underlying mechanistic features.
- Centre for Medical Biotechnology, Faculty of Biology, University Duisburg-Essen, Essen, Germany.
Organizational Affiliation: