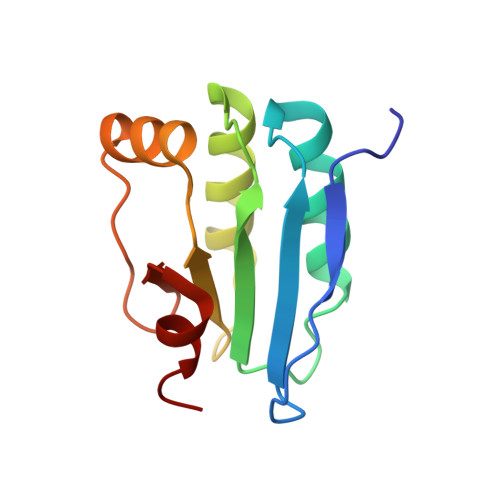
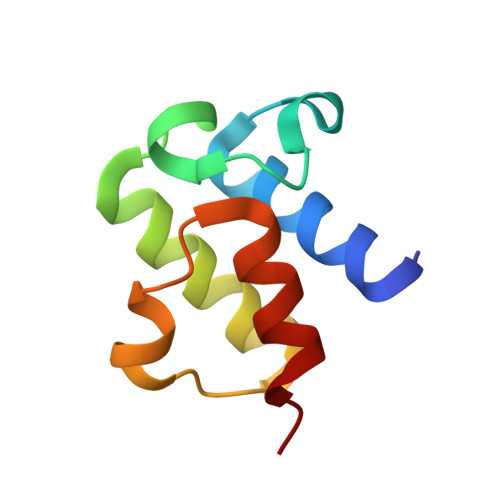
Structure of a SLC26 Anion Transporter STAS Domain in Complex with Acyl Carrier Protein: Implications for E. coli YchM in Fatty Acid Metabolism.
Babu, M., Greenblatt, J.F., Emili, A., Strynadka, N.C., Reithmeier, R.A., Moraes, T.F.(2010) Structure 18: 1450-1462
- PubMed: 21070944
- DOI: https://doi.org/10.1016/j.str.2010.08.015
- Primary Citation of Related Structures:
3NY7 - PubMed Abstract:
Escherichia coli YchM is a member of the SLC26 (SulP) family of anion transporters with an N-terminal membrane domain and a C-terminal cytoplasmic STAS domain. Mutations in human members of the SLC26 family, including their STAS domain, are linked to a number of inherited diseases. Herein, we describe the high-resolution crystal structure of the STAS domain from E. coli YchM isolated in complex with acyl-carrier protein (ACP), an essential component of the fatty acid biosynthesis (FAB) pathway. A genome-wide genetic interaction screen showed that a ychM null mutation is synthetically lethal with mutant alleles of genes (fabBDHGAI) involved in FAB. Endogenous YchM also copurified with proteins involved in fatty acid metabolism. Furthermore, a deletion strain lacking ychM showed altered cellular bicarbonate incorporation in the presence of NaCl and impaired growth at alkaline pH. Thus, identification of the STAS-ACP complex suggests that YchM sequesters ACP to the bacterial membrane linking bicarbonate transport with fatty acid metabolism.
- Banting and Best Department of Medical Research, Terrence Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, 160 College Street, Toronto, ON M5S3E1, Canada.
Organizational Affiliation: