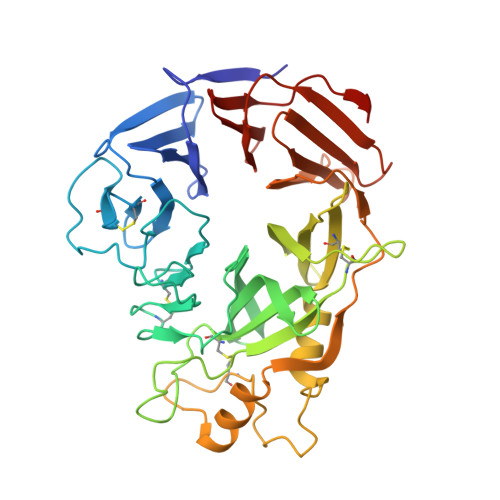
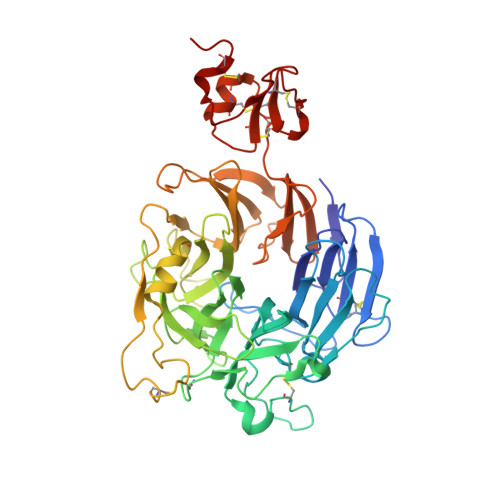
Structural Basis of Semaphorin-Plexin Recognition and Viral Mimicry from Sema7A and A39R Complexes with PlexinC1.
Liu, H., Juo, Z.S., Shim, A.H., Focia, P.J., Chen, X., Garcia, K.C., He, X.(2010) Cell 142: 749-761
- PubMed: 20727575
- DOI: https://doi.org/10.1016/j.cell.2010.07.040
- Primary Citation of Related Structures:
3NVN, 3NVQ, 3NVX - PubMed Abstract:
Repulsive signaling by Semaphorins and Plexins is crucial for the development and homeostasis of the nervous, immune, and cardiovascular systems. Sema7A acts as both an immune and a neural Semaphorin through PlexinC1, and A39R is a Sema7A mimic secreted by smallpox virus. We report the structures of Sema7A and A39R complexed with the Semaphorin-binding module of PlexinC1. Both structures show two PlexinC1 molecules symmetrically bridged by Semaphorin dimers, in which the Semaphorin and PlexinC1 beta propellers interact in an edge-on, orthogonal orientation. Both binding interfaces are dominated by the insertion of the Semaphorin's 4c-4d loop into a deep groove in blade 3 of the PlexinC1 propeller. A39R appears to achieve Sema7A mimicry by preserving key Plexin-binding determinants seen in the mammalian Sema7A complex that have evolved to achieve higher affinity binding to the host-derived PlexinC1. The complex structures support a conserved Semaphorin-Plexin recognition mode and suggest that Plexins are activated by dimerization.
- Department of Molecular Pharmacology and Biological Chemistry, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Organizational Affiliation: