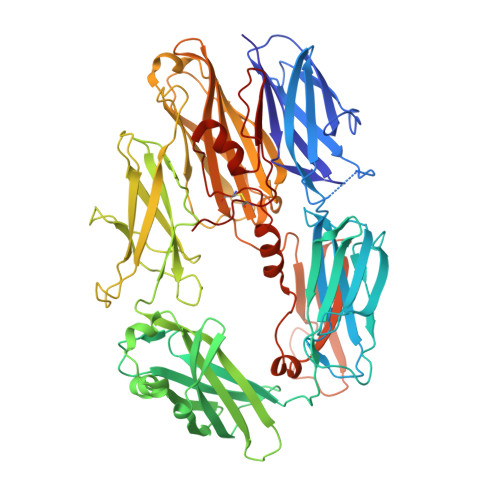
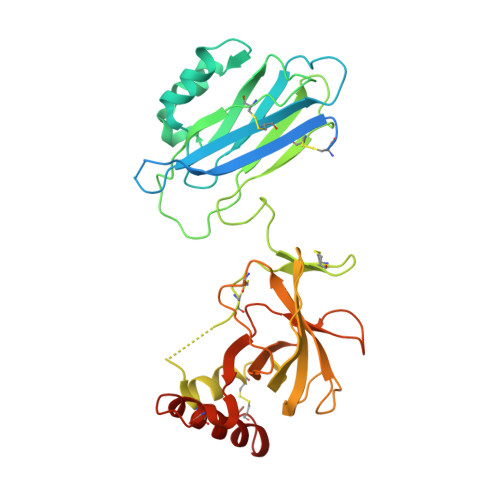
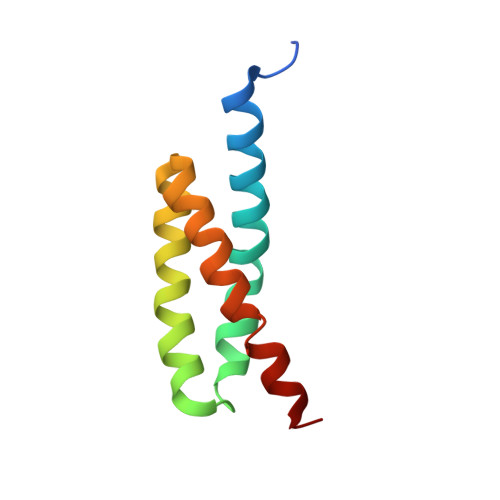
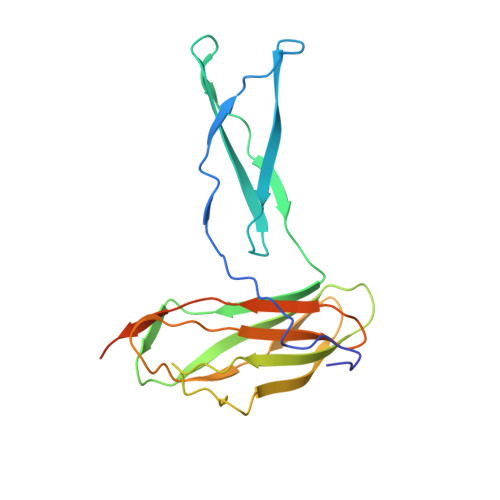
Molecular Basis for Complement Recognition and Inhibition Determined by Crystallographic Studies of the Staphylococcal Complement Inhibitor (SCIN) Bound to C3c and C3b.
Garcia, B.L., Ramyar, K.X., Tzekou, A., Ricklin, D., McWhorter, W.J., Lambris, J.D., Geisbrecht, B.V.(2010) J Mol Biology 402: 17-29
- PubMed: 20654625
- DOI: https://doi.org/10.1016/j.jmb.2010.07.029
- Primary Citation of Related Structures:
3L3O, 3L5N, 3NMS, 3OHX - PubMed Abstract:
The human complement system plays an essential role in innate and adaptive immunity by marking and eliminating microbial intruders. Activation of complement on foreign surfaces results in proteolytic cleavage of complement component 3 (C3) into the potent opsonin C3b, which triggers a variety of immune responses and participates in a self-amplification loop mediated by a multi-protein assembly known as the C3 convertase. The human pathogen Staphylococcus aureus has evolved a sophisticated and potent complement evasion strategy, which is predicated upon an arsenal of potent inhibitory proteins. One of these, the staphylococcal complement inhibitor (SCIN), acts at the level of the C3 convertase (C3bBb) and impairs downstream complement function by trapping the convertase in a stable but inactive state. Previously, we have shown that SCIN binds C3b directly and competitively inhibits binding of human factor H and, to a lesser degree, that of factor B to C3b. Here, we report the co-crystal structures of SCIN bound to C3b and C3c at 7.5 and 3.5 A limiting resolution, respectively, and show that SCIN binds a critical functional area on C3b. Most significantly, the SCIN binding site sterically occludes the binding sites of both factor H and factor B. Our results give insight into SCIN binding to activated derivatives of C3, explain how SCIN can recognize C3b in the absence of other complement components, and provide a structural basis for the competitive C3b-binding properties of SCIN. In the future, this may suggest templates for the design of novel complement inhibitors based upon the SCIN structure.
- Division of Cell Biology and Biophysics, School of Biological Sciences, University of Missouri-Kansas City, 5100 Rockhill Road, Kansas City, MO 64110, USA.
Organizational Affiliation: