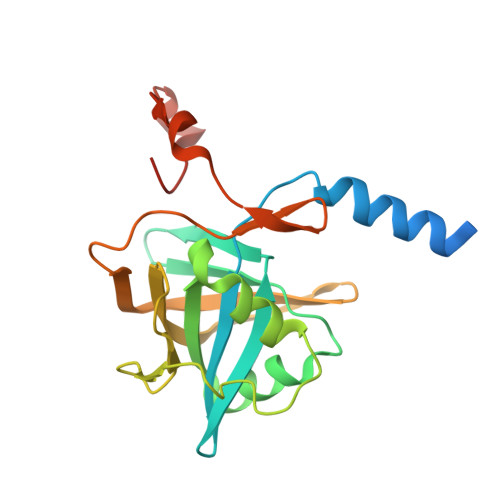
Structure of nitrilotriacetate monooxygenase component B from Mycobacterium thermoresistibile.
Zhang, Y., Edwards, T.E., Begley, D.W., Abramov, A., Thompkins, K.B., Ferrell, M., Guo, W.J., Phan, I., Olsen, C., Napuli, A., Sankaran, B., Stacy, R., Van Voorhis, W.C., Stewart, L.J., Myler, P.J.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1100-1105
- PubMed: 21904057
- DOI: https://doi.org/10.1107/S1744309111012541
- Primary Citation of Related Structures:
3NFW - PubMed Abstract:
Mycobacterium tuberculosis belongs to a large family of soil bacteria which can degrade a remarkably broad range of organic compounds and utilize them as carbon, nitrogen and energy sources. It has been proposed that a variety of mycobacteria can subsist on alternative carbon sources during latency within an infected human host, with the help of enzymes such as nitrilotriacetate monooxygenase (NTA-Mo). NTA-Mo is a member of a class of enzymes which consist of two components: A and B. While component A has monooxygenase activity and is responsible for the oxidation of the substrate, component B consumes cofactor to generate reduced flavin mononucleotide, which is required for component A activity. NTA-MoB from M. thermoresistibile, a rare but infectious close relative of M. tuberculosis which can thrive at elevated temperatures, has been expressed, purified and crystallized. The 1.6 Å resolution crystal structure of component B of NTA-Mo presented here is one of the first crystal structures determined from the organism M. thermoresistibile. The NTA-MoB crystal structure reveals a homodimer with the characteristic split-barrel motif typical of flavin reductases. Surprisingly, NTA-MoB from M. thermoresistibile contains a C-terminal tail that is highly conserved among mycobacterial orthologs and resides in the active site of the other protomer. Based on the structure, the C-terminal tail may modulate NTA-MoB activity in mycobacteria by blocking the binding of flavins and NADH.
- Seattle Structural Genomics Centre for Infectious Disease (SSGCID), USA. sunny.zhang@seattlebiomed.org
Organizational Affiliation: