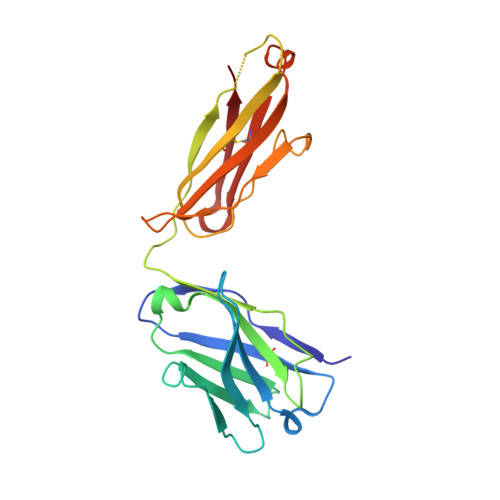
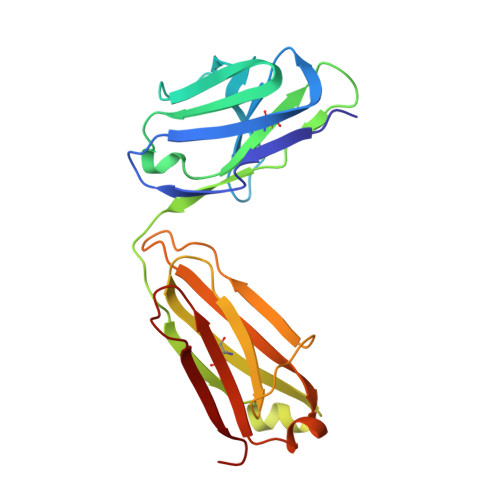
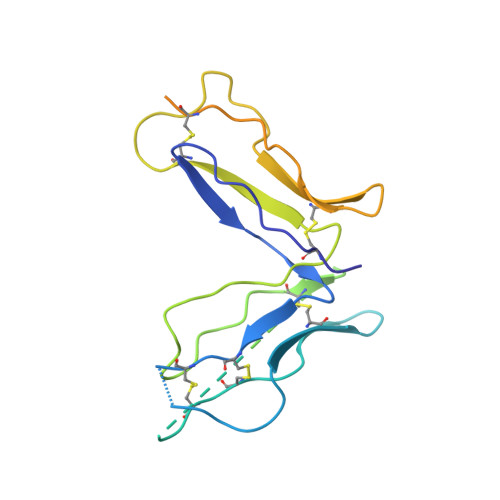
Structural basis of immunosuppression by the therapeutic antibody daclizumab
Yang, H., Wang, J., Du, J., Zhong, C., Zhang, D., Guo, H., Guo, Y., Ding, J.(2010) Cell Res 20: 1361-1371
- PubMed: 20820193
- DOI: https://doi.org/10.1038/cr.2010.130
- Primary Citation of Related Structures:
3NFP, 3NFS - PubMed Abstract:
Interleukin-2 (IL)-2 signaling plays a pivotal role in the activation of immune responses, and drugs that block this pathway have been shown to be effective for the immunosuppression in patients with organ transplantation to alleviate/eliminate allograft rejection. The first humanized monoclonal antibody (mAb) daclizumab falls into this category and shows high specificity and affinity against a key component of the IL-2 receptor complex, namely IL-2Rα. To reveal the molecular mechanism of the inhibition of the IL-2 signaling pathway by daclizumab, we determined the crystal structures of the daclizumab Fab in free form and in complex with the IL-2Rα ectodomain at 2.6 and 2.8 Å resolution, respectively. The daclizumab Fab adopts a similar conformation in the presence or absence of the IL-2Rα ectodomain. The antigen-binding site of daclizumab is mainly composed of five complementarity determining regions (CDRs) that form a large positively charged surface depression and two flanking patches that are generally hydrophobic. The conformational epitope consists of several discontinuous segments of the IL-2Rα ectodomain, a large portion of which overlaps with the regions that interact with IL-2, suggesting that the binding of daclizumab to IL-2Rα would prevent the IL-2 binding to IL-2Rα and the subsequent formation of the IL-2/IL-2Rαβγ(c) complex, and therefore block the IL-2 signaling pathway. These results also have implications for the design and development of improved mAb drugs targeting IL-2Rα.
- State Key Laboratory of Molecular Biology and Research Center for Structural Biology, Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: