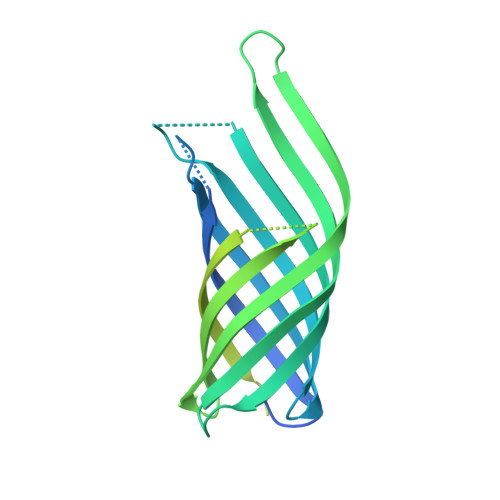
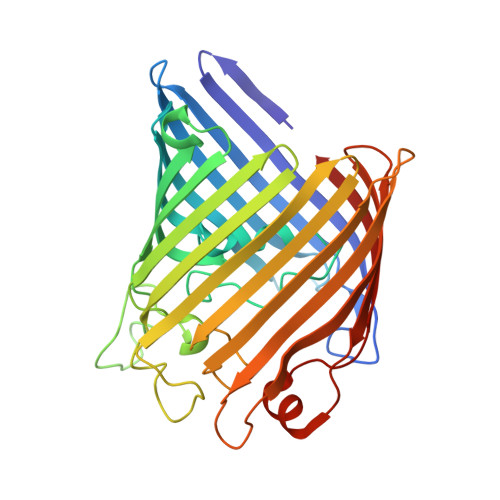
The host outer membrane proteins OmpA and OmpC are associated with the Shigella phage Sf6 virion.
Zhao, H., Sequeira, R.D., Galeva, N.A., Tang, L.(2011) Virology 409: 319-327
- PubMed: 21071053
- DOI: https://doi.org/10.1016/j.virol.2010.10.030
- Primary Citation of Related Structures:
3NB3 - PubMed Abstract:
Assembly of dsDNA bacteriophage is a precisely programmed process. Potential roles of host cell components in phage assembly haven't been well understood. It was previously reported that two unidentified proteins were present in bacteriophage Sf6 virion (Casjens et al, 2004, J.Mol.Biol. 339, 379-394, Fig. 2A). Using tandem mass spectrometry, we have identified the two proteins as outer membrane proteins (OMPs) OmpA and OmpC from its host Shigella flexneri. The transmission electron cryo-microscopy structure of Sf6 shows significant density at specific sites at the phage capsid inner surface. This density fit well with the characteristic beta-barrel domains of OMPs, thus may be due to the two host proteins. Locations of this density suggest a role in Sf6 morphogenesis reminiscent of phage-encoded cementing proteins. These data indicate a new, OMP-related phage:host linkage, adding to previous knowledge that some lambdoid bacteriophage genomes contain OmpC-like genes that express phage-encoded porins in the lysogenic state.
- Department of Molecular Biosciences, University of Kansas, 1200 Sunnyside Avenue, Lawrence, KS 66045, USA. zhaohy@ku.edu
Organizational Affiliation: