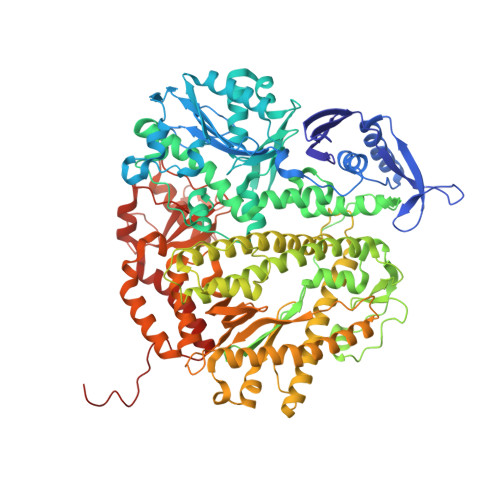
Substitution of Ala for Tyr567 in RB69 DNA Polymerase Allows dAMP and dGMP To Be Inserted opposite Guanidinohydantoin .
Beckman, J., Wang, M., Blaha, G., Wang, J., Konigsberg, W.H.(2010) Biochemistry 49: 8554-8563
- PubMed: 20795733
- DOI: https://doi.org/10.1021/bi100913v
- Primary Citation of Related Structures:
3NAE - PubMed Abstract:
Continuous oxidative damage inflicted on DNA produces 7,8-dihydro-8-oxoguanine (8-oxoG), a commonly occurring lesion that can potentially cause cancer by producing G → T transversions during DNA replication. Mild oxidation of 8-oxoG leads to the formation of hydantoins, specifically guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp), which are 100% mutagenic because they encode almost exclusively the insertion of dAMP and dGMP (encoding G → T and G → C transversions, respectively). The wild-type (wt) pol α family DNA polymerase from bacteriophage RB69 (RB69pol) inserts dAMP and dGMP with low efficiency when situated opposite Gh. In contrast, the RB69pol Y567A mutant inserts both of these dNMPs opposite Gh with >100-fold higher efficiency than wt. We now report the crystal structure of the "closed" preinsertion complex for the Y567A mutant with dATP opposite a templating Gh (R-configuration) in a 13/18mer primer-template (P/T) at 2.0 Å resolution. The structure data reveal that the Y to A substitution provides the nascent base pair binding pocket (NBP) with the flexibility to accommodate Gh by allowing G568 to move in the major-to-minor groove direction of the P/T. Thus, Gh is rejected as a templating base by wt RB69pol because G568 is inflexible, preventing Gh from pairing with the incoming dATP or dGTP base.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8024, USA.
Organizational Affiliation: