Structural basis of ligand selectivity in human CRFR1 and CRFR2 alpha extracellular domain
Pal, K., Swaminathan, K., Pioszak, A.A., Xu, H.E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Maltose binding protein-CRFR2 alpha extracellular domain | 482 | Homo sapiens | Mutation(s): 0 Gene Names: Human CRFR2 alpha | 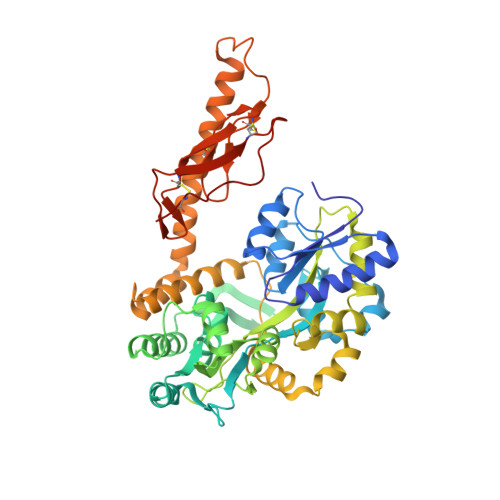 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0AEX9 (Escherichia coli (strain K12)) Explore P0AEX9 Go to UniProtKB: P0AEX9 | |||||
Find proteins for Q13324 (Homo sapiens) Explore Q13324 Go to UniProtKB: Q13324 | |||||
PHAROS: Q13324 GTEx: ENSG00000106113 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q13324P0AEX9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Urocortin-2 | 17 | N/A | Mutation(s): 1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q96RP3 (Homo sapiens) Explore Q96RP3 Go to UniProtKB: Q96RP3 | |||||
PHAROS: Q96RP3 GTEx: ENSG00000145040 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q96RP3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900001 Query on PRD_900001 | G, H, I, J | alpha-maltose | Oligosaccharide / Nutrient |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 54.311 | α = 90 |
| b = 212.068 | β = 104.54 |
| c = 107.319 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| MAR345dtb | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASES | phasing |