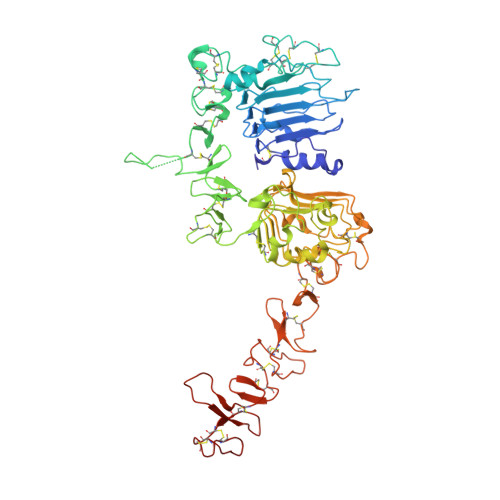
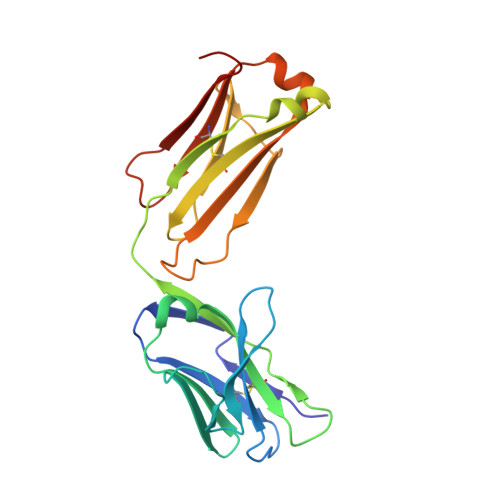
Structure of the complex between HER2 and an antibody paratope formed by side chains from tryptophan and serine.
Fisher, R.D., Ultsch, M., Lingel, A., Schaefer, G., Shao, L., Birtalan, S., Sidhu, S.S., Eigenbrot, C.(2010) J Mol Biology 402: 217-229
- PubMed: 20654626
- DOI: https://doi.org/10.1016/j.jmb.2010.07.027
- Primary Citation of Related Structures:
3N85 - PubMed Abstract:
Engineered antibody paratopes with limited sequence diversity permit assessment of the roles played by different amino acid side chains in creating the high-affinity, high-specificity interactions characteristic of antibodies. We describe a paratope raised against the human ErbB family member HER2, using a binary diversity tryptophan/serine library displayed on phage. Fab37 binds to the extracellular domain of HER2 with sub-nanomolar affinity. An X-ray structure at 3.2 A resolution reveals a contact paratope composed almost entirely of tryptophan and serine residues. Mutagenesis experiments reveal which of these side chains are more important for direct antigen interactions and which are more important for conformational flexibility. The crystal lattice contains an unprecedented trimeric arrangement of HER2 closely related to previously observed homodimers of the related epidermal growth factor receptor.
- Department of Protein Engineering, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: