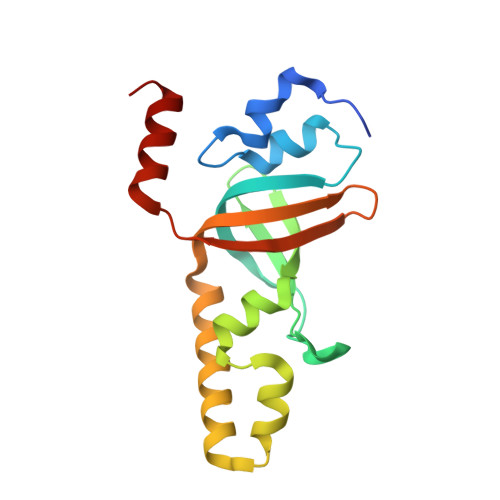
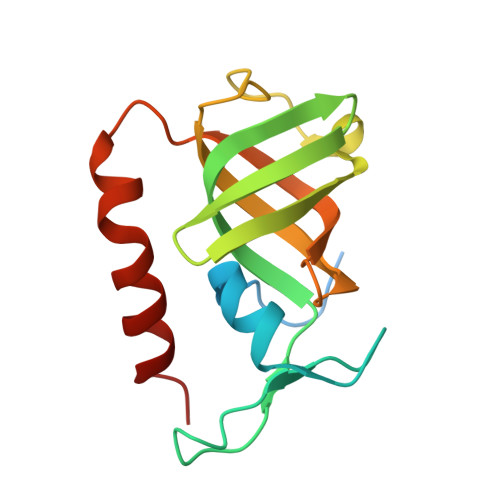
Structure and cellular roles of the RMI core complex from the bloom syndrome dissolvasome.
Hoadley, K.A., Xu, D., Xue, Y., Satyshur, K.A., Wang, W., Keck, J.L.(2010) Structure 18: 1149-1158
- PubMed: 20826341
- DOI: https://doi.org/10.1016/j.str.2010.06.009
- Primary Citation of Related Structures:
3MXN - PubMed Abstract:
BLM, the protein product of the gene mutated in Bloom syndrome, is one of five human RecQ helicases. It functions to separate double Holliday junction DNA without genetic exchange as a component of the "dissolvasome," which also includes topoisomerase IIIα and the RMI (RecQ-mediated genome instability) subcomplex (RMI1 and RMI2). We describe the crystal structure of the RMI core complex, comprising RMI2 and the C-terminal OB domain of RMI1. The overall RMI core structure strongly resembles two-thirds of the trimerization core of the eukaryotic single-stranded DNA-binding protein, Replication Protein A. Immunoprecipitation experiments with RMI2 variants confirm key interactions that stabilize the RMI core interface. Disruption of this interface leads to a dramatic increase in cellular sister chromatid exchange events similar to that seen in BLM-deficient cells. The RMI core interface is therefore crucial for BLM dissolvasome assembly and may have additional cellular roles as a docking hub for other proteins.
- Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, 550 Medical Science Center, 1300 University Avenue, Madison, WI 53706-1532, USA.
Organizational Affiliation: